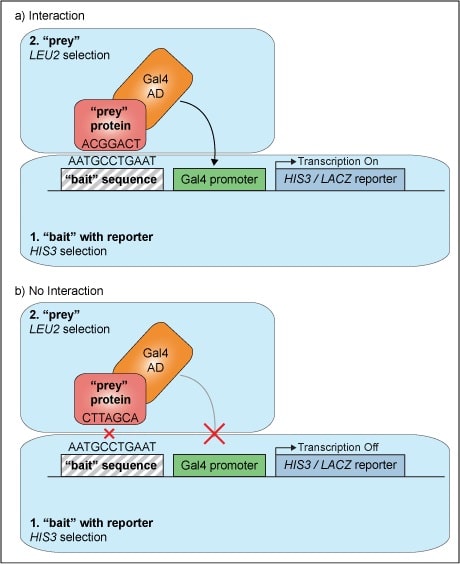
An Introduction To ChIP-seq
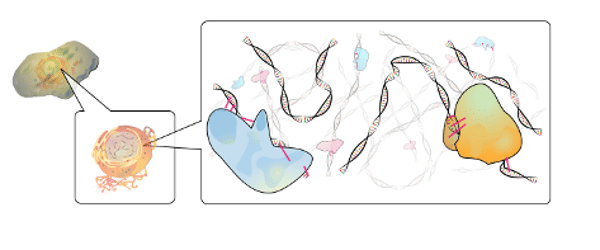
…more commonly ChIP is interrogated with microarrays (ChIP-chip) or next-generation sequencing (ChIP–seq). How does ChIP-chip work? ChIP-on-chip, or ChIP-chip, combines chromatin immunoprecipitation with microarray (chip) analysis. In 2001, Jason Lieb…