If you are regularly doing ChIP-qPCR, ChIP-RNAseq or luciferase reporter assays to measure protein-DNA interactions, then this article is for you!
ChIP experiments can tell you what DNA sequences your protein binds, and luciferase reporter assays predict whether your protein functionally binds a specific promoter to activate transcription – but a yeast one-hybrid (Y1H) assay flips these around and tells you what proteins bind a given DNA sequence in vivo. Interested? Read on to find out more!
So what is a Y1H assay?
Y1H relies on the general principles of the yeast two-hybrid assay. In short, mammalian proteins are exogenously expressed in yeast and their interactions in vivo are measured by the downstream activation of reporter gene constructs. The main difference between Y1H and Y2H is in the interactions that are being measured – put simply, Y2H measures protein-protein interactions, while Y1H measures protein-DNA interactions.
How does it work?
In Y2H experiments, the close physical interaction between two proteins (the bait and the prey) brings together the DNA-binding and activation domains of a transcription factor. This forms a functional transcriptional unit that turns on expression of a reporter gene.
In a Y1H system, detection is based on the interaction of a single protein (the prey) with a bait DNA sequence positioned upstream of a reporter gene. Importantly, the prey protein is fused to a transcriptional activation domain. Positive protein-DNA interactions bring the fusion activation domain into close proximity with the promoter element, thus activating downstream transcription of the reporter gene as seen in the figure below.
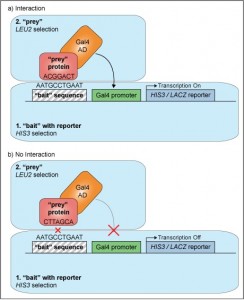
You will need:
1) A DNA bait sequence. This can be a defined promoter sequence, another defined cis-acting sequence, regulatory sequences from exons, introns, 5’ or 3’-noncoding regions, or even centromere or telomere sequences.
2) Bait plasmid. This also doubles as your reporter plasmid. Unlike Y2H, which uses a standard reporter construct for all bait:prey interactions, Y1H reporter constructs are specially designed for your experiment to contain the reporter gene (commonly HIS3, a histidine biosynthesis gene, or LacZ, which encodes for bacterial enzyme ?-galactosidase) under the control of a non-endogenous promoter which lies immediately downstream of your DNA bait sequence. This construct is commonly delivered using a special plasmid that is able to reliably and stably integrate the sequence by site-specific mutagenesis into a nonessential part of the yeast genome (e.g. pINT). Successful transformation is then tested using histidine-deficient selection media.
Expert tip: this selection step is important for Y1H as it indicates the level of background signal. Some DNA bait sequences may be bound and activated by yeast transcription factors, resulting in leaky expression of HIS3 even in the absence of a prey plasmid. This can be reduced (using competitive inhibitors of the HIS3 enzyme such as 3-AT) to improve your signal:noise ratio, but it is crucial to optimise this with histidine dropout medium so that you can correctly identify a positive Y1H result later on.
3) Prey plasmid (or plasmids). Much like in a Y2H experiment, your prey plasmid will encode for a protein-of-interest, called the “prey”, as well as a selection marker, e.g. a leucine biosynthesis gene, which will allow the yeast to grow in medium lacking leucine. However, a big difference in Y1H is that your prey protein is expressed as a fusion protein with a strong constitutive trans-activation domain (e.g. Gal4, LexA or VP16) that is able to bind and activate the promoter region just upstream of the reporter gene. This is to ensure you achieve robust transcriptional activation of the HIS3 reporter gene, regardless of whether the prey protein has any strong intrinsic activation function.
You’ll also need all the other reagents and equipment you would use for a standard Y2H, such as genetically modified yeast, selection markers, and yeast growth media. Check out our Overview of Yeast Two-Hybrid (Y2H) Screening for more details and some help on how to design your experiment.
Why choose a Y1H assay?
Although it shares some similarities with other DNA-protein detection approaches, a Y1H assay has some clear advantages:
- Unlike luciferase reporter assays, Y1H is able to detect protein-DNA interactions that are not actually activating transcription. Fusing your protein(s) of interest to a strong activation domain allows Y1H to detect a variety of DNA-binding proteins, including those that do not directly function in transcription, e.g. replication proteins, DNA repair proteins, and repressor proteins.
- Y1H is compatible with many existing libraries. Most Y1H experiments can use hybrid prey libraries that have been constructed for Y2H applications, e.g. Gal4p- or LexA-based protein libraries can also be used for screening against various DNA baits in Y1H.
- Y1H can detect isoform-specific interactions. You can design your prey plasmids to encode specific protein isoforms, allowing detection of highly specific interactions. In contrast, ChIP-based approaches rely on sensitive and specific antibody binding to your protein of interest, and these are rarely able to be achieved on an isoform-specific level.
Like most yeast-based screening approaches, Y1H has some disadvantages too:
- Y1H doesn’t provide any information about the functional consequences of a DNA-protein interaction.
- The rates of false-positive results are high, as either endogenous yeast transcription factors or prey proteins that can bind Gal4 promoters can activate transcription of the reporter gene without interacting with the bait DNA sequence.
- Expression of your prey protein fused to an activation domain can result in improper protein folding, resulting in steric hindrance and loss of interactions, and/or failure to correctly localise to the nucleus.
Has a Y1H assay worked for you? Let us know in the comments below how you have used this technique in your research!







