The epigenome has been in the research spotlight, and for good reason. Not only has it been associated with the developmental stages of an organism, but epigenetic alterations lead to disorders and have been linked to many human diseases. So, the question stands: what exactly is an epigenome?
What Is the Epigenome?
Simply put, the epigenome is every chemical alteration in DNA or histones (proteins that bind to the DNA) that affects gene expression; however, it does not alter the DNA sequence. This means that sequencing a genome is only half of the story, as some secrets are hiding away in the epigenome.
How is this possible? One of the most important (and studied) epigenetic modifications is the methylation of cytosine on the CpG dinucleotide. About 70%-80% of CpG dinucleotides are methylated in human cells. However, CpG islands (sequences with a high percentage of CpG dinucleotides, situated in the 5´-end regulatory regions near the transcription start site of most human genes) are found to be unmethylated in healthy cells. Abnormal methylation of these sites may lead to unregulated gene expression.
DNA methylation has been linked to embryonic development, and evidence proposes that methylation influences gene expression. Different methylation patterns have been found on different cell types, suggesting that it plays a critical role in silencing genes and, therefore, in cell differentiation. It is important to remember that methylation patterns are inherited and maintained during normal cell division, meaning that every daughter cell has the same methylation pattern.
Why Is It Important to Study Methylation?
Knowledge is power. And even though there is still a lot to learn about the epigenome and the role of methylation on the homeostasis of the cell, it is safe to say that it plays a big role in disease and development. Indeed, abnormal patterns of methylation have been associated with serious human disorders, like developmental diseases such as Prader-Willi syndrome, autoimmune diseases, and cancer, among others.
Abnormal methylation patterns found in tumor cells indicates the prominent role that methylation plays in tumorigenesis. The genome of cancer cells is widely hypomethylated, which leads to chromosomic instability. Interestingly, hypermethylation is found on target genes in cancer cells (silencing genes related to suppressing tumors), which permits metastasis of the tumor. The hypermethylation of these genes may be a disease biomarker and used for an early diagnosis.
The epigenome, unlike the DNA sequence, is not set in stone. This means that epigenetic alterations are reversible. This opens a new field of therapeutic medicine, as we can use methylation to stop the epigenetic modifications that lead to disease, and we can increase the expression of a desired trait to fight disorders. When studied thoroughly, it may become a great therapeutic advantage!
How Can We Study Methylation?
Methylation of DNA can be studied through different methods. However, current approaches for genome-wide DNA methylation analysis are based on three principles:
- Bisulfite conversion
- Antibody or affinity-based enrichment
- Methyl-sensitive restriction enzymes
With the advent of Next Generation Sequencing (NGS), these principles have been integrated in its platforms, to get a large set of data regarding methylation in a short time. We are going to discuss some of the techniques that use the NGS platform to study methylation (Table 1)!
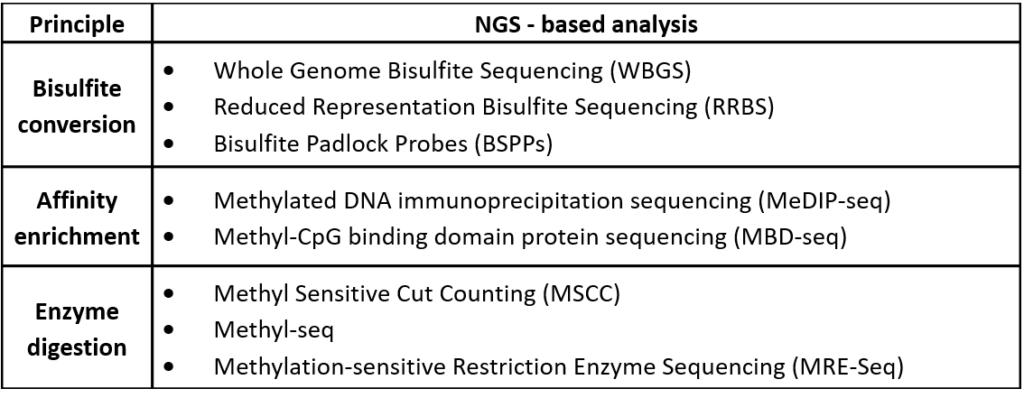
Table 1 – Different techniques used to study DNA methylation. Adapted from Laird, Peter W. “Principles and challenges of genome-wide DNA methylation analysis.” Nature Reviews Genetics 11.3 (2010): 191.
- Bisulfite conversion
Bisulfite treatment modifies every non-methylated cytosine into uracil through deamination. However, methylated cytosines are resistant to this modification, and therefore don’t change. So we can discern between a methylated cytosine and an unmethylated one. When reading a sequence treated with bisulfite, every cytosine we find is from a methylated spot.
- Whole Genome Bisulfite Sequencing
Whole Genome Bisulfite Sequencing (WGBS) is a great technique that shows the methylation status of the whole genome, by using bisulfite conversion. However, since it aims to find the whole methylome, it becomes costly due to the mandatory sequencing depth.
- Reduced Representation Bisulfite Sequencing
Reduced Representation Bisulfite Sequencing (RRBS) is a technique that also relies on bisulfite conversion to distinguish between methylated and unmethylated DNA. However, it also relies on digestion of DNA by enzymes insensitive to methylation. MspI is one of the most commonly used endonucleases in this technique, as it recognizes the site CmCGG. This step leads to a decrease in cost as it selects fragments with CpG. Therefore, it gives us fragments on a silver plate that can be methylated. Because this enzyme is not sensitive to methylation it will always digest the DNA on the specific sequence, even if it is methylated. This technique selects fragments of the genome that could be methylated and thus contrasts with the WBGS approach. However, MspI may not cleave every site susceptible to methylation, and therefore some genomic regions may have low coverage, or may not be represented at all.
- Bisulfite Padlock Probes (BSPPs)
This technique combines bisulfite conversion with padlock probes. These probes have a shared linker sequence and are hybridized to the DNA (previously treated with bisulfite, so that un-methylated cytosine deaminates into uracil). This product is then circularized and PCR amplified. The beauty of all of this is that the probes contain a restriction site for an endonuclease, so that every fragment will have the same size, making it ready for NGS.
This technique improves the yield result in the enrichment step by combining the annealing specificity of the probes and the amplification with universal primers leads to an even representation of the fragments. Nevertheless, because capture occurs after bisulfite conversion, there is a concern that it may interfere with the capture step and therefore distort the methylation measurements.
Antibody or affinity-based enrichment
Antibody and affinity-based enrichment techniques use molecules that bind specifically to methylated DNA. This specificity allows for enrichment, as all the unbound DNA and therefore unmethylated DNA can be washed away, leaving only the methylated DNA to be sequenced. The techniques based on this principle allow for a quick and effective genome-wide valuation of methylation but do not give info on individual CpG dinucleotides.
- Methylated DNA immunoprecipitation sequencing (MeDIP-seq)
MeDIP-seq relies on immunoprecipitation to enrich the methylated fragments of DNA. In this technique, the monoclonal anti-methyl-cytosine antibody binds specifically to methylated areas of the genomic DNA, resulting in the removal of all unmethylated DNA. Thus, all that will remain is a library of fragments of methylated DNA, ready to be sequenced.
- Methyl-CpG binding domain protein sequencing (MBD-seq)
The MBD family of proteins contains a methyl-binding domain (MBD) and these proteins have an affinity to symmetrically methylated sites. MBD proteins bind specifically to symmetrical methylated sites. Consequently, the MBD-seq technique allows one to discard unmethylated DNA, and to maintain only the methylated genomic DNA that will remain as a library to be sequenced.
Methylation-sensitive restriction enzymes
Endonuclease digestion is a powerful tool in molecular biology. In fact, this was the first tactic used to learn more about the epigenome and DNA methylation.
Restriction enzymes cleave the DNA at precise restriction sites, specific to each enzyme. Some enzymes are sensitive to methylated regions, and will provide a different cutting pattern for the DNA, depending on the methylation state of the restriction site. There are several techniques that are translated to NGS. The downfall of these techniques lies with the endonuclease´s restriction site – the methylated sites can be hidden away in other sequences.
- Methyl-Sensitive Cut Counting (MSCC)
This technique uses HpaII, a methylation-sensitive restriction enzyme, to cut genomic DNA. The action of this enzyme is prevented when its recognition sequence CmCGG is methylated. Hence, by using this enzyme, all fragments obtained will be from unmethylated sites. The unmethylated fragments will be the library to be sequenced, and can then infer which sites are methylated.
- Methyl-seq
This technique, much like MSCC, also uses the endonuclease HpaII to cleave the unmethylated genomic DNA. However, it differs from MSCC by using the isoschizomer MspI as a control. MspI is insensitive to methylation, and therefore cleaves its restriction site (CmCGG) regardless of the methylation state. After digestion, the genomic DNA is ligated to NGS adapters, selected by size, and ready to be sequenced. Therefore, all the reads corresponding to MspI cleavage that are not present in the library obtained by HpaII cleavage are fragments derived from methylated sites.
- Methylation-sensitive Restriction Enzyme Sequencing (MRE-Seq)
MRE-seq also is based on the use of endonucleases sensitive to methylation. However, this technique stands out because, unlike the previous techniques, it uses various methylation-sensitive restriction enzymes to obtain more information. By using different methylation-sensitive restriction enzymes, with different restriction sites, we are studying more methylated sites because we can see more sites. After digestion, the library can be sequenced.
Concluding Remarks
The epigenome is an exciting topic for further understanding human disease and development. While there is a lot we have learned, there is still far more to be uncovered.
Tell us, what’s your favorite method to study the epigenome? Which one do you prefer?
References
- Laird, Peter W. “Principles and challenges of genome-wide DNA methylation analysis.” Nature Reviews Genetics 11.3 (2010): 191.
- Robertson, Keith D. “DNA methylation and human disease.” Nature Reviews Genetics 6.8 (2005): 597.