A Protein Renaturation Toolkit: 21 Tips and Tricks for Refolding Proteins

Expressing your protein of interest but not sure if it’s properly folded? Read on to discover advice and tips for refolding proteins.
The Problem of Inclusion Bodies
Bacterial protein expression is a great way to get large quantities of your protein for assays, structural studies, and more. But convenience often turns to frustration when you’ve expressed your recombinant protein a few times and it refuses to come out in your purification and instead remains stuck in inclusion bodies (IBs). This happens when your protein doesn’t fold properly during expression, a problem that can be fixed through protein renaturation.
In this article, we’re going to discuss ways you can fix the IB problem and check that your protein is correctly folded and functional.
If you’re looking for tips on getting bacteria to express your target protein in the first place, consult our articles on points to remember before expressing proteins in bacterial systems and optimizing bacterial protein expression.
Why Would I Need to Renature My Protein?
The majority of experiments on a protein—e.g., finding its structure by crystallography or other methods, tracking its localization in cells, or testing it on animal models—require it to be folded into its native conformation, which can entail structural formations such as:
- secondary structures including alpha-helices and beta-sheets;
- tertiary structural motifs such as leucine zippers, zinc fingers, and disulfide bonds;
- and quaternary structures such as dimers, tetramers, and larger complexes.
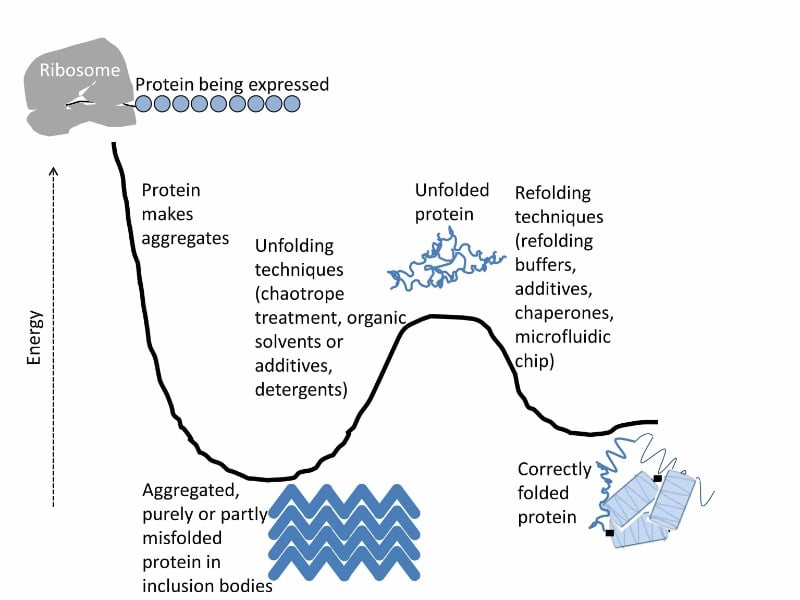
Your protein should be solvated in your solution or properly bound to a solid surface. If the protein is misfolded, aggregating with other molecules of itself, and crashing out of its solution, its native structure is likely inaccessible to its current energy state (Figure 1), without which most experiments would lack biological relevance.
The Disadvantages of Bacterial Expression
Expressing a recombinant protein in bacterial systems, despite having practical advantages including affordability and usually high yield, has disadvantages when the target is a eukaryotic protein. Let’s look at these in a bit more detail.
Problematic Post-Translational Modifications
Bacterial cells lack the machinery to add post-translational modifications (PTMs) to proteins, a hallmark of eukaryotic cell function. A lack of PTMs not only leads to restrictions in the protein’s physiologically relevant functionality when assessed in experiments, but also can be prohibitive to correct folding into its native structure, which is supposed to occur on expression.
Insufficient Chaperones
In addition, while bacteria do express chaperones, which guide proteins toward proper folding, expression protocols generally involve overexpression to produce the target protein in large amounts, rapidly exhausting bacterial cells’ natural chaperone machinery.
As a result, the literature is replete with descriptions of and how-tos about bacterially expressed recombinant eukaryotic proteins that end up in IBs, which are proteinaceous deposits inside or outside host cells in which expressed protein has aggregated and crashed out of solution. [1-5] In this IB state, the protein is insoluble and can’t be used as-is for experiments.
What Does It Take to Successfully Refold a Protein?
Owing to extensive variations in properties such as size, charge, and hydrophobicity, different proteins respond differently to the same treatments; thus, there is no one-size-fits-all approach. But don’t worry, we’re going to walk you through a series of approaches to refolding proteins, so you’ll be sure to find one that suits your experiment.
Tip 1: First, Rescue the Protein from Inclusion Bodies
Dissolving a protein from an IB by most conventional protocols involves treating it with a concentrated denaturant or chaotrope, usually urea or guanidine hydrochloride at a concentration of 6–8 M. [2,3] Alternatively, you could use a strong detergent such as sodium dodecyl sulfate (SDS) (2% w/v). [5]
Denaturant is usually present as part of a physiological-pH buffer, which is used to wash IBs repeatedly until they dissolve. Washing generally involves sonicating IBs in the buffer or placing the IB–buffer mixture in a tube on a rocker, alternating with centrifugation steps. You can run Western blots on your pellets and supernatants from your centrifugations to check that your protein successfully came out in the supernatant.
At the end of this solubilization step, the protein should be solvated. For it to be usable, however, it will have to be renatured because denatured protein is completely or almost completely unfolded and prevented by the chaotrope or detergent from forming its native structure. The denaturant will have to be removed by dilution, dialysis, or a less conventional method (see below).
Tip 2: Find a Protocol for Refolding Proteins
A good early step in figuring out your protocol would be to look up your target protein in the REFOLD database, [6–8] which provides thousands of protocols (2060 refolding methods, at the time of writing) for renaturing different proteins.
If your protein is not listed there, or it is but the methods suggested don’t work out for you, don’t panic! We still have plenty of tips and tricks for you to try, so read on.
Tip 3: Try Dilution or Dialysis
The primary conventional methods for solubilizing IBs are dilution and dialysis. Dilution simply means that the protein in the denaturant or chaotrope is diluted in a refolding buffer. Dialysis involves placing the denatured protein in a semi-permeable bag or tube with semi-permeable membrane and in turn floating that in an (at least) 1000-fold volume of refolding buffer.
The principle is that the refolding buffer gradually replaces the denaturant solution inside the tube or bag, while the pore size of the dialysis membrane is selected to be too small to permit leakage of the protein itself into the exterior buffer.
These methods have disadvantages. Following dilution, the resulting large volume of highly diluted protein—generally 100- to 1000-fold diluted from its original concentration in the denaturant—may not be useful in experiments, and much of the protein may still aggregate during the dilution process as denaturant becomes more dilute while the protein itself becomes more dilute. [9]
Dialysis may also entail the denaturant concentration decreasing too rapidly to continue preventing protein aggregation. [9]
Striking the balance between diluting or replacing the denaturant gradually enough and promoting native folding with refolding buffer efficiently enough depends on how your particular protein responds to your dilution or dialysis conditions.
Unconventional Techniques for Better Results
For refolding proteins that are particularly tricky, you may need to look beyond dilution and dialysis.
Tip 4: Immobilization and Refolding Proteins on a Gradient
Immobilizing a protein on a chromatography column and then diluting it in refolding buffer on a gradient prevents aggregation in several cases. [10]
Tip 5: Used Detergent? Precipitate It Out!
If you used SDS as a denaturant instead of urea or guanidine hydrochloride (see Tip 1 above), dilution and dialysis aren’t going to cut it. SDS is notoriously stubborn to remove.
Biochemists He and Ohnishi [5] have recently purified ulvan lyase protein by successfully precipitating out SDS (which they’d used as a denaturant) by adding 400 mM KCl and incubating the mixture at 4°C overnight. A reverse-phase column in high-performance liquid chromatography (HPLC) can also remove SDS. [5]
Tip 6: Buy a Refolding Kit
Choosing the right refolding buffer, column, and size of dialysis or dilution step can be like looking for a needle in a haystack if you don’t know where to start.
Fortunately, to optimize conditions, kits are now available that allow you to screen for the right refolding buffers and columns via a matrix system that bypasses much of the extensive time it would take to design and carry out your own troubleshooting experiments from scratch.
Pierce™ Protein Refolding Kit and Athena Sciences’ QuickFold™ Protein Refolding Kit are examples of commercial options designed to screen your conditions. Sometimes a lyophilization step is necessary to maintain the protein in the refolded state until it is ready for use, or to rehydrate it in a reaction buffer that differs from the refolding buffer.
Tip 7: Use a Microfluidic Chip
Another tool worth trying is microfluidic chips. As denatured protein flows on the chip from the central stream to laminar flow, it encounters one or more junctions through which it passes from denaturing solution to refolding buffer. [9] The more junctions in the chip design, the more steps from buffer to buffer, and therefore the more gradual the change in buffer, lowering the risk of aggregation compared with classical one-step dialysis or dilution. [9]
Tip 8: Additives!
One or more chemical additives can be the key to refolding your protein. The amino acids arginine and proline, as well as the non-canonical amino acids arginineamide and glycineamide, have been observed to inhibit aggregation, thus promoting the refolding process. [9]
Urea and guanidinium hydrochloride, usually used at high concentrations to denature proteins as described above, can at low concentrations inhibit aggregation and promote refolding. [9] Glycerol, polyethylene glycol, and sugars such as sucrose and trehalose have worked to stabilize some protein structures during refolding, [9] as have mild detergents such as cholate, Triton X-100, or 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS). [11]
Tip 9: Bring a Chaperone
Alternatively, you may need to recreate what host bacterial cells have failed to do when expressing your protein: use chaperones to guide the protein into its correct structure. Cyclodextrins have proven useful as artificial chaperones for this technique, as they interact with target proteins via capture of the protein in a hydrophobic cavity and weak interaction between the cyclodextrin’s hydrophilic groups and those of the protein. [9]
Tip 10: Solubilize Without a Denaturant
Many proteins form IBs that have been found to contain an equilibrated mixture of misfolded and correctly folded protein. [12] For such proteins, an alternative technique to denaturing and then renaturing protein from IBs is to solubilize the correctly folded protein from IBs without a denaturation step.
Some solubilizing agents [such as sarcosyl and, at low concentrations (5%), dimethyl sulfoxide (DMSO) and n-propanol] are entirely non-denaturing and bypass the need for refolding. [12]
Treating IBs with high-pH buffer (>12), high concentrations (6 M) of organic solvents ([e.g. beta-mercaptoethanol (BME)], or applying high pressure (2–4 kbar) would solubilize but somewhat disrupt without totally denaturing the structure of the target protein.
This would require refolding proteins afterwards but with a much lower loss of protein to aggregation than the denaturation method. [12] (Beware of using BME, however, to solubilize a protein that is reliant on its disulfide bonds for stability and function, such as ribonucleases, which are irreversibly denatured by BME. [13])
Tip 11: Mind Your C’s
Cysteine enables a protein to form disulfide bonds, which need to be considered during solubilization, denaturation, and renaturation steps. Chances are your cysteine-containing protein formed incorrect disulfide bonds while misfolded in IBs.
You’ll need to release those bonds by including a strong reducing agent such as dithiothreitol (DTT) in the denaturing solution. Then, the refolding buffer should contain an optimized balance of redox agents, [10] such as a mix of reduced and oxidized glutathione in a ratio ranging from 10:1 to 1:1, or else a cysteine–cystine couple at a 10:1 ratio.
This will establish a redox equilibrium that will promote the formation of correct disulfide bonds as your protein refolds while inhibiting incorrect disulfide bond formation. As with overall refolding, you may need a troubleshooting step to adjust and readjust these amounts according to their effect on your protein.
Techniques for Bypassing Inclusion Bodies Altogether
If the optimal tools for correctly refolding your protein continue to prove elusive, at some point you’ll want to circumvent the need for denaturing/renaturing by avoiding IB formation in the first place.
Tip 12: Add a Tag!
A lot of proteins have been successfully expressed with a thioredoxin, glutathione S-transferase, or maltose-binding protein tag and easily dissolved in lysis buffer without forming IBs. Chances are, however, that your experiment will require you to work with the protein without the tag.
Still, cleaving off the tag as a downstream step makes it easier to control the state of the protein’s folding—typically by adjusting the buffer pH, including organic solvents, or diluting the protein [14,15]—than when the protein is still in the bacterial cells.
Tip 13: Remove Problematic Residues
Perhaps only a small cluster of amino acids, or maybe just one amino acid, is causing your target protein to aggregate during expression.
An online tool such as that developed by Hamodrakas et al. [16] can help you predict polypeptide regions that trigger IB formation. If these residues aren’t crucial for your experiment, you could try expressing the protein without them.
Tip 14: Divide and Conquer
It could also be an option to express one or more domains of your protein but not the whole protein. Some experiments work well, or even better, using only the relevant protein fragment.
Large proteins can be divided up into modules—generally according to functionality, hydrophobic and hydrophilic amino acid-dominated regions, or encoding exon—and insoluble modules can be left out of the expression vector (see Loftus et al. [17] and Gross et al. [18], for example).
Tip 15: Go Eukaryotic
Finally, you may consider avoiding IB formation by not using bacterial cells for your protein expression. Insect cells are a widely optimized eukaryotic system for expression that can make target proteins at a large scale and treat them with the proper chaperones and post-translational modifications.
Tip 16: Ditch the Cell
Another possibility is not to use cells at all. A cell-free expression system would produce proteins at a lower concentration, making IB formation unlikely, and, due to containing only the macromolecules you actually put into the system, afford the biochemical flexibility to add co-solutes or make other modifications to optimize preservation of the native structure of your target protein.
Did Protein Refolding Work?
Once you’ve obtained a good yield of your protein in your buffer of choice, you’ll still want to check the structure to make sure it’s folded into its native conformation.
If part of your experiment is to research a protein’s structure because it isn’t known in the first place, that check is built into your work, although it’s tricky to tell whether the structure is correct if you don’t know what structure you’re looking for!
Tip 17: Use Circular Dichroism to Check for Refolding
Circular dichroism (CD) is an ideal way to evaluate solubility and secondary structure, as it reveals spectra with signature peaks according to whether a protein sample contains disordered regions, beta-sheets, beta-turns, alpha-helices, and/or polyproline helices. [19]
You might know a related protein that has a likely similar structure to that of your new protein, or you could use software such as PSIPRED or AlphaFold [20,21] to predict the structure of your target protein. If the CD spectra match up somewhere in the ballpark of those similarities or predictions, it’s safe to say your protein is properly folded.
In addition, if the protein is aggregating, it will likely show inconsistent spectra on measuring the CD of different aliquots of it, or even give a low CD signal owing to poor solvation.
Tip 18: Try Size-Exclusion Chromatography
If your protein is misfolded and aggregating inappropriately, it should have a larger bulk mass than in its desired native state. This can be assessed using size-exclusion chromatography (SEC) [4] or non-denaturing polyacrylamide gel electrophoresis (PAGE), also known as a native gel, to ascertain that the protein is running at the correct molecular weight and cross-sectional size.
Again, if you have a similar protein in the lab with better-understood behavior, you can run it through the SEC column or in a neighboring lane in the gel as a positive control.
Tip 19: Check for Disulfide Bonds Using SDS-PAGE or Non-Denaturing PAGE
Gels can also enable you to check for disulfide bonds. If the expected bonds are intermolecular, simply performing SDS-PAGE with and without a strong reducing agent such as BME, [13] which would break all disulfide bonds, would show the protein running more slowly in the absence of the agent due to Cys–Cys bridges between the molecules.
For intramolecular disulfide bonds, running your protein in a native gel with and without BME would be more helpful, as changes in size beyond those caused by molecular weight can be viewed in this way and can be compared to assess whether the desired disulfide bonds have formed.
Tip 20: Find Out if it’s Functional
One more way to test your protein for refolding is to check its functionality. Healthy cells tend to dispose of protein that isn’t properly folded. Therefore, radioactively or fluorescently labeling your protein and tracking its stability in cell culture by performing pulse–chase or bleach–chase experiments, respectively, could be a way to evaluate its refolding.
Tip 21: Monitor Your Protein’s Interactions
Does your protein form a complex with another protein, or is it an enzyme? Santos et al. [4] assessed the protein–protein interactions of XfPal, a protein they had renatured after expression, by measuring its complex formation with peptidoglycan. In their case, the complex caused XfPal to leave the soluble fraction for the insoluble fraction after a set of centrifugations were performed.
Another nifty way to check for protein–protein interactions is co-immunoprecipitation followed by Western blot to detect the proteins you expect to have formed a complex. If your target protein is an enzyme, functionality could be measured via a kinetic assay such as the one described in this working with enzymes article.
Refolding Proteins in a Nutshell
We appreciate that there’s a lot of information to take in here, so we’ve created a handy table to outline the various steps and tools for protein renaturation covered in this article.
Table 1. Steps for successful protein renaturation
Desired step | Recommended technique(s) or additive(s) |
Hunt for protocols | REFOLD database, shop around for refolding kits (e.g., Pierce™, FoldIt™ by Hampton Research, QuickFold™ by Athena Sciences) |
Denature proteins misfolded in inclusion bodies (IBs) | Urea or guanidinium hydrochloride (6–8 M), SDS (2%) |
Solubilize proteins from IBs without completely denaturing them | High pH (>12) and BME or n-propanol (6 M) with urea (2 M) |
High pressure (2–4 kbar), as with French press | |
Refold proteins directly from IBs | Sarcosyl, DMSO (5%), n-propanol |
Renature proteins after denaturation | Techniques
|
Dilution, dialysis, chromatography column, precipitate out SDS with KCl or use reverse-phase HPLC, refolding kits, lyophilization to change buffers, microfluidic chips | |
Additives
| |
Urea or guanidinium hydrochloride (1–2 M) | |
Arginine, proline, arginineamide, glycineamide | |
Glycerol, polyethylene glycol, sucrose, trehalose | |
Cholate, Triton X-100, CHAPS | |
Cyclodextrins (use as refolding chaperones) | |
Promote correct formation of disulfide bonds | DTT during denaturation, followed by reduced and oxidized glutathione during renaturation at 1:1–10:1 ratio or cysteine–cystine at 10:1 |
Bypass IB formation | Inclusion of a tag (thioredoxin, glutathione S-transferase, or maltose-binding protein), followed by tag removal (enzymatic cleavage while adjusting pH, including organic solvents, and/or diluting protein) |
Eliminate one or more amino acids that promote aggregation | |
Express only one domain or group of domains needed of target protein | |
Express in a eukaryotic system (e.g., insect cells) | |
Express in a cell-free system | |
Analyze protein for proper refolding | Circular dichroism, while comparing measured structure with structure predicted by software (e.g., PSIPRED, AlphaFold) or structure of homolog |
Size exclusion chromatography, non-denaturing gel, using BME to check for disulfide bonds | |
Pulse–chase/bleach–chase, protein–protein interactions (co-immunoprecipitation, fractionation via ultracentrifugation), enzymatic assay |
It may be a long and winding road to properly folded recombinant protein, but we hope this toolkit of tips and tricks will help you get there. When the time comes for your experiments, it will all be worth it! Have we missed any tips for refolding proteins? Let us know in the comments below.
References
- Bell S, Hansen S, Buchner J. Refolding and structural characterization of the human p53 tumor suppressor protein. Biophys. Chem., May 2, 2002;doi: https://doi.org/10.1016/s0301-4622(02)00011-x
- Asano R, Kudo T, Makabe K, Tsumoto K, Kumagai I. Antitumor activity of interleukin-21 prepared by novel refolding procedure from inclusion bodies expressed in Escherichia coli. FEBS Lett., Sep. 25, 2002;doi: https://doi.org/10.1016/S0014-5793(02)03254-4
- Petrlova J, Hong H-S, Bricarello D, Harishchandra G, Lorigan G, Jin L-W, Voss JC. A differential association of Apolipoprotein E isoforms with the A-beta oligomer in solution. Proteins, Feb. 2011;doi: https://doi.org/10.1002/prot.22891
- Santos CA, Beloti LL, Toledo MAS, Crucello A, Favaro MTP, Mendes JS, Santiago AS, Azzoni AR, Souza AP. A novel protein refolding protocol for the solubilization and purification of recombinant peptidoglycan-associated lipoprotein from Xylella fastidiosa overexpressed in Escherichia coli. Protein Expr. Purif., Jan. 28, 2012;doi: https://doi.org/10.1016/j.pep.2012.01.010
- He C & Ohnishi K. Efficient renaturation of inclusion body proteins denatured by SDS. Biochem. Biophys. Res Comm., July 3, 2017;doi: https://dx.doi.org/10.1016/j.bbrc.2017.07.003
- REFOLDdb. Accessed Nov. 13, 2021.
- Chow MK, Amin AA, Fulton KF, Fernando T, Kamau L, Batty C, Louca M, Ho S, Whisstock JC, Bottomley SP, Buckle AM. The REFOLD database: a tool for the optimization of protein expression and refolding. Nucleic Acids Res., Jan. 1, 2006;doi: https://doi.org/10.1093/nar/gkj080
- Phan J, Yamout N, Schmidberger J, Bottomley SP, Buckle AM. Refolding Your Protein with a Little Help from REFOLD. Methods Mol. Biol., May 30, 2011;doi: https://doi.org/10.1007/978-1-60327-223-0_4
- Yamaguchi H & Miyazaki M. Refolding Techniques for Recovering Biologically Active Recombinant Proteins from Inclusion Bodies. Biomolecules, Feb. 20, 2014;doi: https://doi.org/10.3390/biom4010235
- Middelberg APJ. Preparative protein refolding. TRENDS Biotechnol., Aug. 19, 2002;doi: https://doi.org/10.1016/s0167-7799(02)02047-4
- Johnson M. Detergents: Triton X-100, Tween-20, and More. Labome: The world of laboratories. Accessed Nov. 14, 2021.
- Singh A, Upadhyay V, Upadhyay AK, Singh SM, Panda AK. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microbial Cell Factories, March 25, 2015;doi: https://doi.org/10.1186/s12934-015-0222-8
- Wikipedia. 2-Mercaptoethanol. Accessed Nov. 19, 2021.
- Shen, A. Simplified Protein Purification Using an Autoprocessing, Inducible Enzyme Tag. Methods Mol. Biol., May 29, 2014;doi: https://doi.org/10.1007/978-1-4939-1034-2_5
- Pandey NK, Isas JM, Rawat A, Lee RV, Langen J, Pandey P, Langen R. The 17-residue-long N terminus in huntingtin controls stepwise aggregation in solution and on membranes via different mechanisms. J. Biol. Chem., Feb. 16, 2018;doi: https://doi.org/10.1074/jbc.M117.813667
- Tsolis AC, Papandreou NC, Iconomidou VA, Hamodrakas SJ. A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins. PLoS One, Jan. 10, 2013;doi: https://doi.org/10.1371/journal.pone.0054175
- Loftus SR, Walker D, Maté MJ, Bonsor DA, James R, Moore GR, Kleanthous C. Competitive recruitment of the periplasmic translocation portal TolB by a natively disordered domain of colicin E9. Proc. Natl. Acad. Sci., Aug. 15, 2006;doi: https://doi.org/10.1073/pnas.0603433103
- Gross GG, Junge JA, Mora RJ, Kwon H-B, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GCR, McGee AW, Sabatini BL, Roberts RW, Arnold DA. Recombinant Probes for Visualizing Endogenous Synaptic Proteins in Living Neurons. Neuron, June 19, 2013;doi: https://dx.doi.org/10.1016/j.neuron.2013.04.017
- Circular Dichroism and the Conformational Analysis of Biomolecules. Ed.: GD Fasman. New York: Plenum Press, 1996;doi: https://doi.org/10.1007/978-1-4757-2508-7
- PSIPRED Workbench. Accessed Nov. 18, 2021.
- AlphaFold Protein Structure Database. Accessed Nov. 18, 2021.