Homology Terminology: Never Say the Wrong Word Again
I’ve noticed that there exists some ambiguity about the different terms relating to homology. I’ll try to break them down here, with significant help from Genome Biology.
Join Us
Sign up for our feature-packed newsletter today to ensure you get the latest expert help and advice to level up your lab work.
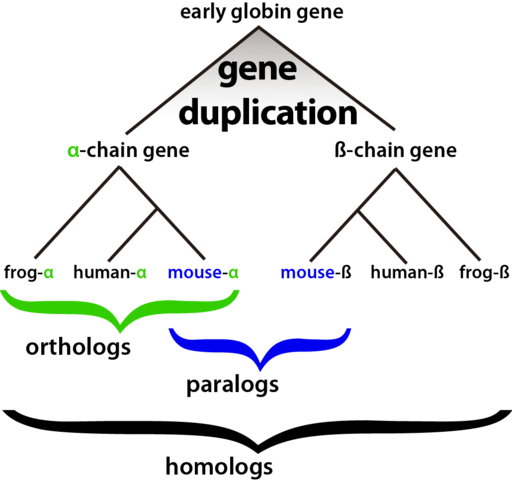
I’ve noticed that there exists some ambiguity about the different terms relating to homology. I’ll try to break them down here, with significant help from Genome Biology.

Now you’ve got great sequencing results, thanks to Nick’s article on improving sequencing results. Now what? Well now you need some software (preferably free) to analyze your data. BioEdit is a good option. But what I have to offer today is a much lighter and equally handy tool. It’s called Artemis and was developed by…

In the sci-fi novel Terminal World by Alistair Reynolds, a planet consists of zones with defined characteristics of matter interactions on a subatomic level. These conditions permit different levels of technology sophistication in various zones. For example, in the “Steamville zone” nothing more complicated than steam engines works – electronic schemes fuse irreversibly. Something like…
A disruptive sequencing technology Every new generation, a new concept is born and can completely reshape the landscape of biomedical research. Nanopore sequencing technology, although still at its infancy, is beginning to look like a “game-changer.” It’s a revolutionary concept in sequencing in which strands of nucleic acids are fed through a tiny pore (nanopore)…
Ion Torrent technology, when it was introduced in 2010, was one of several machines that promised to revolutionize genetics. These were benchtop machines that showed their prowess in quickly sequencing smaller exomes and other DNA samples (about 10-20 million bases per run, compared to Illumina HiSeq, which could read 250 billion bases in a run)….

So, you’re sitting there with your list of significant SNPs, thinking, “what do I do now”? Hopefully this article can point you in the right direction! So far, you will have extracted genomic DNA from your organism of interest, sourced the SNP chips required, and had the DNA run on these chips. The chips will…

Genomics, transcriptomics, proteomics, metabolomics – words that in 2015 sound very familiar even to a freshman in any biology field. Although most have heard those words before, I keep encountering students or even post-graduates who find it difficult to explain what they are. So, to make things easier here is a peek behind the curtains…
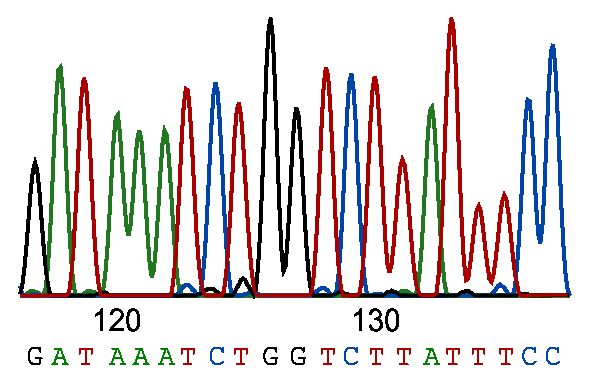
I’ve never run a sequencing gel in my life, but people around me did, and they spent a lot of time on getting it just right. Although the principle described by Sanger in 1975 sounds straightforward (1), sequencing gels are very long and very thin – less than a millimeter thick! They were easy to…

Welcome to the magical world of systematics! Looking for a way to produce a phylogenetic tree that’s a step above the default options, time efficient, not too program heavy and avoids using command line programs? Although there are more rigorous analyses that strict systematists perform, for your purposes, the following should suffice. 1. Data selection…

The development of CRISPR/Cas9 technology has made it relatively straightforward to selectively edit genomes and has revolutionized the way in which we approach biological questions. CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats and in simple terms, this technique allows you to direct a nuclease to cut at a specific site in the genome of interest. The…

Anchored multiplex PCR (AMP) is a powerful method for amplifying minuscule amount of nucleic acids. Combined with Next Generation sequencing, AMP just might be what you need to identify genetic mutations.

Genomic Science has come a long way since the early days of Sanger sequencing in the 1970’s. Today, there are jazzy new sequencing technologies that include fragment analysis, epigenetic sequencing, RNA/transcriptome sequencing and Next Generation Sequencing (NGS). Increasingly these technologies are becoming more accessible, but they still require highly specialized (read: expensive) equipment. Unless your…
Recently, I have witnessed the uprising of various next generation sequencing (NGS) platforms and it’s quite interesting because each platform uses a different method. Previously, I’ve written about the exciting possibility of nanopore sequencing—a new sequencing technology based on the “signature” electrical currents generated as a single strand of DNA passes through the nanopore. The…

While it is true that there are some useful websites like SNPedia, or NCBI that can help you find rs codes for genetic variants, sometimes you need that info coming straight from the oven – particularly when you want to look at atypic SNPs or substitutions that have not been validated. So, in this post I…
Welcome to the last article in this series! Last, but by no means least, we will look at the importance of plant miRNAs and how they differ from their animal counterparts. When/How Were Plant miRNAs Discovered? Plant miRNAs were first described in 2002, a decade after the seminal miRNA study in the nematode C. elegans…
Size selection is a critical step in NGS pipelines, but may be most challenging for studies of small RNAs. The concept behind size selection is simple: separate a sheared DNA or cDNA sample by fragment size, and then use the resulting sizes to remove unwanted fragments. This is a tried-and-true way to get rid of…
It took scientists a little while to warm up to long-read sequencing, but now you couldn’t pry most of them away from their sequencers with a crowbar. Long reads — we’re talking 10,000 bases and more — provide a level of contiguity and completeness in genome assemblies that simply isn’t possible with short-read sequencers. They…
The age of sequencing is undoubtedly upon us. From improving cancer diagnostics to pinning down elephant poaching hotspots, DNA sequencing is revolutionizing the world around us from the ground up. The latest video from Thermo Fisher Scientific’s “Behind the Bench” blog, 10 moments in DNA sequencing gives fascinating insights into the amazing advances being made…
Of all the sample prep steps necessary for next generation sequencing, DNA size selection may have the greatest impact on quality of results. After all, ineffective sizing can waste sequencing capacity on low molecular weight material such as adapter-dimers or primer-dimers, while imprecise sizing can prevent bioinformaticians from producing accurate assemblies. High-quality size selection can…

Do you want to know more about the story behind some of today’s big developments in sequencing technology? Illumina recently started a video series called “Adventures in Genomics” that introduces the people working on methods and applications, making big waves in our understanding of science, biology, and medicine. The company’s most recent video covers single-cell…
If you’re new to next gen sequencing, figuring out what to do with your results can be a daunting process. Luckily, you’re not alone—plenty of people have been in your shoes, and there is tons of information about data analysis out there. Here are some free resources you can use to get up to speed…

Next gen sequencing is a powerful technique, one that now lies at the heart of many scientific projects. This power comes with some special challenges, however, and by recognizing them you can ensure that your NGS results are robust. No one wants to publish findings that other scientists fail to replicate, but unfortunately it happens…

The recent advancement of next generation sequencing technology and the development of novel gene editing tools, such as CRISPR-Cas9, have revolutionized research in genetics. In this golden era of molecular biology, knowing how to dig and navigate through all the enormous sequence information is an essential skill for most molecular biologists. However, to obtain facile…
You don’t need to be told about how next generation sequencing technologies have revolutionized the way we study the genome and the epigenome. Whether you want to look at transcription (RNA-seq), translation (Ribo-seq) genomes (DNA-seq), interactions of proteins and DNA (ChIP-Seq) or to study epigenetic features such as methylation (whole genome bilsulfite sequencing) there are…

It was not long since the commercialization of NGS (a little more than ten years ago) that scientists went beyond the basics and got creative with the new technology to study much more than just the sequence of DNA. In this article we highlight some of the different NGS technologies and methods available out there….

“Making the simple complicated is commonplace; making the complicated simple, awesomely simple, that’s creativity.” – Charles Mingus Next Generation Sequencing (NGS) technology has boomed in recent years, allowing researchers to probe further into the workings of the genome. According to the theory of simplicity, it is the simple principles on its basis that make…
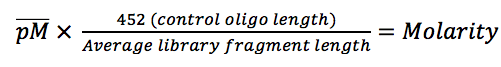
If you want to get the maximum yield and quality from your next-generation sequencing experiment then you are going to need to make sure each of the libraries you produce is carefully quantified ready for pooling and/or loading onto a flow cell. If the quantification goes wrong you’ll get a bad balance of samples within…

In my previous article ‘Choosing a scripting language for next gen sequencing: Python, Perl, and more’ I discussed several of the more common programming languages used for next generation sequencing and things to consider when picking which one to learn. But now that you know WHAT you want to learn, HOW do you go about…

Large amounts of data? Check. Repetitive tasks? Check. If you work with next gen sequencing data, you have probably already realized it’s a good idea to learn a scripting language. But learning a programming language is a major endeavour, and with lots of languages available how do you decide which one to study? And once…

Making a Next Generation Sequencing (NGS) library can seem a bit daunting to the new user, as failures can be expensive. But don’t be put off, as NGS library preparation is relatively simple molecular biology, and can be very easy if you choose to use a commercial kit from one of the many suppliers. Take…

The Rise and Fall of the 454 Sequencer The GS20 454 sequencer, released in 2005, was the first next-generation DNA sequencer to hit the market, and its feats quickly dazzled the scientific community. As new sequencing platforms proliferated, however, many researchers opted for less expensive options and 454 market share fell. About a year ago,…

The completion of the Human Genome Project in 2003 ushered in a new era of rapid, affordable, and accurate genome analysis—called Next Generation Sequencing (NGS). NGS builds upon “first generation sequencing” technologies to yield accurate and cost-effective sequencing results. Fred Sanger sequenced the first whole DNA genome, the virus phage ?X174, in 1977. In that…

What do these three seemingly disparate studies have in common? After publication, the high-profile findings in each one were questioned due to the presence of batch effects. Batch effects are ever-present and insidious in science, so as researchers we need to always be on guard against them. Keep reading for a run-down of how they…

You’re about to start that big project you’ve been dreaming of for years. You’ve identified a potential miracle compound and want to figure out how it affects gene expression. But how are you going to do it: with next gen sequencing or a microarray? Especially if you are new to this area of research, the…
DNA sequencing (PCR, Sanger or next-generation sequencing (NGS)) is a now familiar part of any molecular biology lab. But ‘RNA-seq’, the so-called “Cinderella of genetics”, is now becoming the belle of the ball, providing new insights into this most central molecule of the ‘central dogma’. The many flavors of RNA Whilst genomic DNA is the…

Bisulfite pyrosequencing is becoming a routine technique in molecular biology labs as a method to precisely measure DNA methylation levels right down to the single base. The technique allows for detailed and high resolution analysis of DNA methylation at specific genomic regions. How to detect the 5th base? Methylation of any of the four nucleotides…

Biologists have long appreciated the complexity of genome organization, but until recently lacked the tools to discern the intricacies of this puzzle. Now, thanks to some handy cross-linking, careful amplification, and (of course!) next generation sequencing, teams from Massachusetts are taking us down the rabbit hole, with some surprising findings from Wonderland. Bend Over Backwards…
The Irish Famine (or ‘Great Potato Famine’ if you live outside the Emerald Isle) killed one million people and forced another million to leave the country between 1845 and 1852. It was caused by a blight on the country’s main food stock- the Irish ‘Lumper’ potato. Now, researchers have identified the genome of the blight…

I was first introduced to Conrad Waddington’s epigenetic landscape when reading ‘The epigenetic revolution’, a fantastic introduction to epigenetics, and in my opinion, a must read for anyone who is looking for an entertaining and enjoyable introduction to this fascinating field. In his model, Waddington likens the process of cellular differentiation to a marble, which…

The field of epigenetics is exploding and given the strong links between epigenetic state and disease, the need to study markers like DNA methylation in humans is very relevant. This article outlines some of the main factors you should be taking into account in your study of DNA methylation in human tissues. Here goes: Biological…

The eBook with top tips from our Researcher community.