It’s an all-too-common scenario: You’re reading a hot new paper and, while perusing the figure captions, you come across the phrase “N = 3”. For experiments using animals, this phrase generally means N = 3 animals. But what does “N = 3” mean for cell line experiments? N = 3 cells? Cell culture wells? Independent experiments? Cell lines? Cheeseburgers?
In this article, we demystify the concept of biological replication in cell line experiments and offer practical advice for how you can improve the robustness and generalizability of your cell-line experiments.
Why Use Cell Lines?
Cell lines are often used instead of primary cell cultures and whole organisms for several reasons. They are cheap, readily obtainable, and easy to propagate. They also eliminate the need to use live animals for research because they can be subcultured many times over. Subcultures can be frozen and shared between labs with ease. This feature makes replicating results in cell lines easier than primary cell cultures.
Cell lines also have their fair share of disadvantages. First and foremost, they are often far removed from their in vivo counterparts due to the manipulations used to immortalize them and the absence of tissue-specific factors in commonly used culture media. [1] Cell lines can be contaminated by other cell lines and microbes alike. Also, although the fact that cell lines are generally derived from single biological source helps generate reproducible results, this fact raises an important question about how to interpret these results: is biological replication possible in cell line experiments?
Can Biological Variation Be Tested Using Cell Lines?
Our understanding of the natural world depends on our ability to draw statistical inferences from the experiments we perform. Replication, or the application of a manipulation to multiple biological entities, is critical to this process. [2] The key phrase here is “multiple biological entities”. To understand why robust and generalizable cell line experiments are deceptively hard to design, let’s unpack this concept a bit.
For animal experiments, obtaining and testing multiple biological entities is very straightforward. Simply obtain multiple mice (or whichever animal you are studying) and randomize them into different treatment groups. “N” equals the number of animals you’ve assigned to each group. Easy.
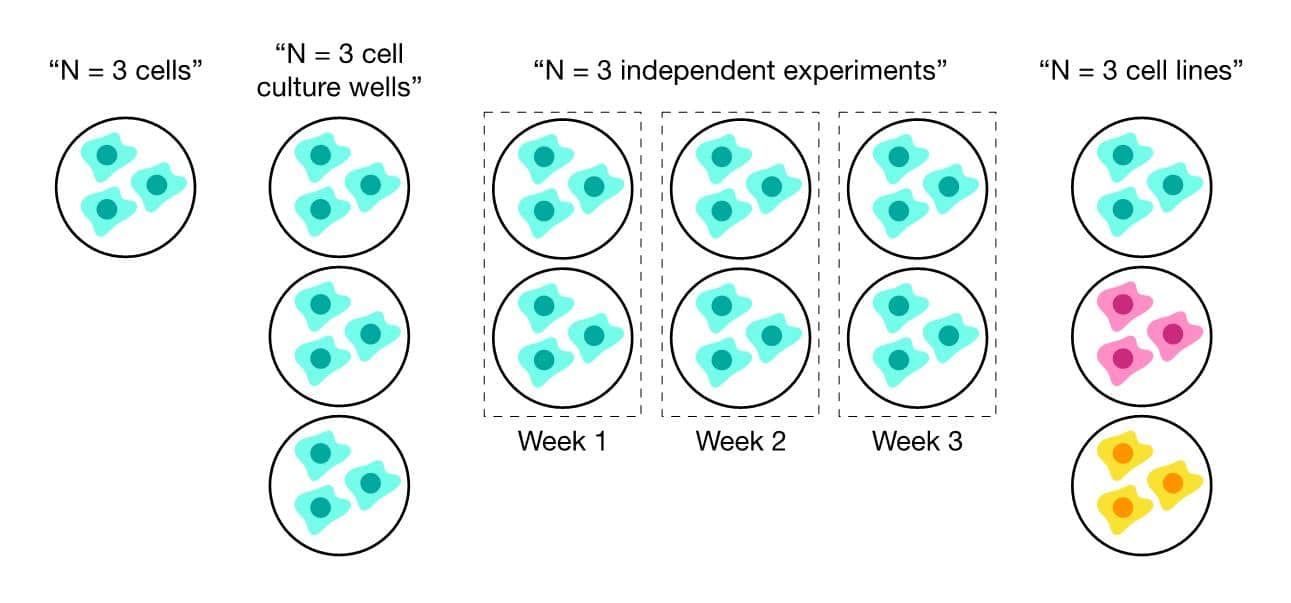
For cell line experiments, the process is much less straightforward. What does “N” mean in this case? Figure 1 illustrates how “N” could mean anything from individual cells to multiple cell lines. [1] However, in the first three cases, only technical replication is being assessed because the illustrated definitions of “N” use a single cell line. Only when multiple cell lines are derived from biologically independent sources (e.g., from different mice) can biological replication truly be tested. [2]
Does this mean your cell line experiments that use a single cell line are worthless? No. But it does mean you should be explicitly clear when defining what “N” means in your experiments.
How to Design Cell Line Experiments
There is ample disagreement in the scientific literature about which definitions of “N” are acceptable for cell line experiments. Although some argue that the “independent experiments” definition is the minimal standard for cell line experiments, [3] others suggest that the “cell culture wells” definition is also acceptable, albeit not very robust. [2] In the latter case, cells are randomized into wells, wells are randomized into treatment groups, and treatments are independently administered to each well. Some might argue that this approach only measures pipetting ability, but a similar argument can be levied against the “independent experiments” definition. Both cases can only test technical variation, whether it’s across wells or the whole experimental protocol.
Using Multiple Monoclonal Cell Lines
A better approach involves isolating multiple monoclonal cell lines from a polyclonal source. In this case, N is defined as “monoclonal cell lines”. Cells within a polyclonal cell line exhibit measurable genetic and phenotypic variation. This definition is often used when characterizing a new cell line, but rarely for run-of-the-mill cell line experiments because most labs do not create their own cell lines. Furthermore, if this definition meets the standard of biological replication is debatable. This is because monoclonal cell lines also exhibit variation due to the immortalization process and cellular senescence with repeated passaging. [4] In other words, monoclonal cell lines do not necessarily represent the natural biological variation we expect to see in the natural world.
Isolating primary cells from multiple animals and subjecting each cell preparation to the same immortalization procedure presents a more robust and generalizable approach. But this is almost never done in practice because it is easier to simply use the primary cells for experiments and avoid immortalization altogether — that is unless you are studying the immortalization process itself.
As you can see, the appropriate definition of “N” for your experiment depends on your experimental goals. The more levels of replications you incorporate in your experiment, the more robust and generalizable it is. For example, a robust experiment might define “N” as “monoclonal cell lines” and would be performed multiple times using multiple cell culture wells for each experimental replicate. This approach is more generalizable than testing a single cell culture well for each monoclonal cell line because it provides more accurate information about the magnitude and variation of the studied effects. [5]
Are Cell Lines Useful?
By now, you might be convinced that cell line experiments are useless. But there are some very productive uses for cell lines. For example, genome-scale screening applications rely on tens of millions of cells to generate hundreds of thousands of gene perturbations for interrogation properly. Cell lines like HEK293T are so amenable to gene transfer that they can be used to reconstruct entire signaling pathways for study. [6] The generation of cell lines can also increase access to biological samples from “exotic” animals such as snakes and bats. In fact, such cell lines are necessary to study pathogens specific to these animals. [7]
At the end of the day, cell lines provide a means to reduce the use of animal subjects in research and improve reproducibility across laboratories. That said, careful planning and documentation are necessary to maximize the generalisability of cell line experiments. Regardless of your cell line application, you can improve the robustness of your data by implementing multiple levels of technical replication in your experimental design, as discussed above.
Want to learn more about cell lines? Check out Bitesize Bio’s articles on the top 5 most common cell lines and how to choose the right one for your experiment.
References
1. Kaur G and Dufour JM (2012) Cell lines. Spermatogenesis 2(1): 1-5. doi: 10.4161/spmg.19885.
2. Lazic SE, et al. (2018) What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol 16(4): e2005282. doi: 10.1371/journal.pbio.2005282.
3. Cumming G, et al. (2007) Error bars in experimental biology. J Cell Biol 177(1): 7-11. doi: 10.1083/jcb.200611141.
4. Geraghty RJ, et al. (2014) Guidelines for the use of cell lines in biomedical research. Br J Cancer 111(6): 1021-1046. doi: 10.1038/bjc.2014.166.
5. Naegle K, et al. (2015) Criteria for biological reproducibility: What does “n” mean? Sci Signal 8(371): fs7. doi: 10.1126/scisignal.aab1125.
6. Shi H, et al. (2016) Reconstruction of the mouse inflammasome system in HEK293T cells. Bio Protoc 6(21): e1986. doi: 10.21769/BioProtoc.1986.
7. Dove A. (2014) The art of culture: Developing cell lines. Science 346(6212): 1013-1015. doi: 10.1126/science.346.6212.1013.