A lot of research experiments require the use of a eukaryotic host as opposed to E. coli due to its greater conformity and suitability in expressing eukaryotic proteins. This is the reason why yeast cells have gained importance as cloning and expression hosts. For protein expression studies to hybrid screens, many applications require insertion and expression of a heterologous gene into the yeast cell. Yeast plasmids have been specifically designed for this purpose1.
Types of Yeast Plasmids
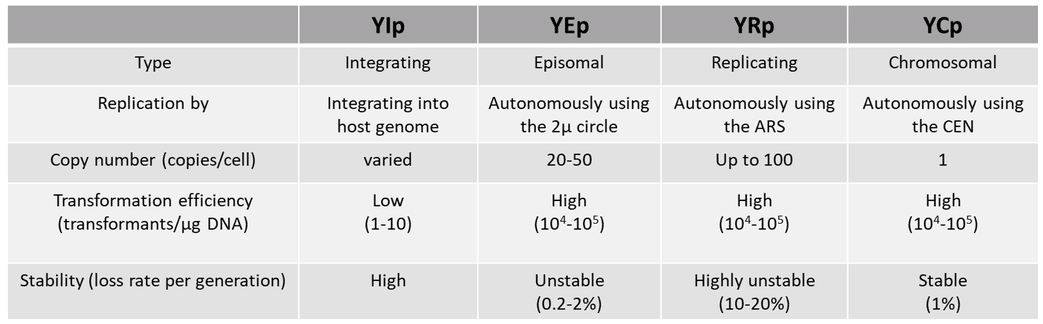
A. YIp (yeast integrating plasmid)
Ylp lacks an origin of replication, and thus the ability to replicate autonomously, so it integrates into the host chromosome for survival and replication. Integration occurs via homologous recombination between a gene on the plasmid and the same gene in the host genomic DNA. Ylp has low transformation frequency (1-10 transformants/µg DNA), which can be enhanced by linearizing the plasmid using restriction enzymes. The copy number of the plasmid depends on the number of genes present in the host genome available for homologous recombination. Copy number can be increased by using a gene in the plasmid that has multiple copies in the genome. The plasmid has high stability but can be lost under lack of selection pressure. This plasmid is used for stable gene integration into the genome or to replace a host gene with another gene or its mutant.
B. YEp (yeast episomal plasmid)
YEp is an extrachromosomal plasmid that replicates autonomously using the 2µ origin. It has high transformation efficiency (104 – 105 transformants/µg DNA) and a high copy number (20-50 copies/cell). It is a stable plasmid (loss rate 0.2 – 2% per generation) and can be used for overexpression studies where high amounts of the recombinant protein are essential.
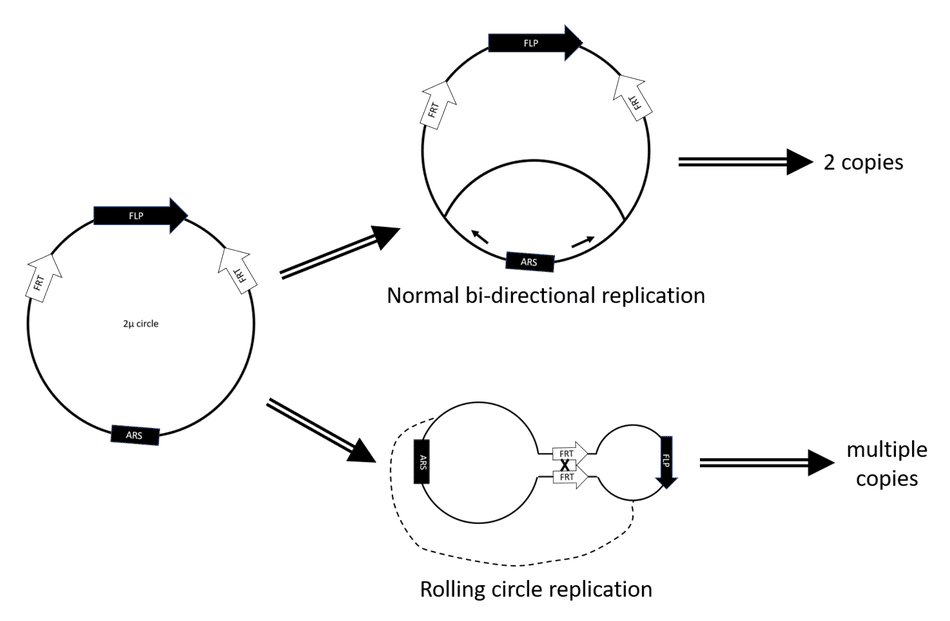
C. YRp (yeast replicating plasmid)
Similar to a YEp, YRp is also an extrachromosomal plasmid that replicates autonomously with the help of a chromosomal element called ARS (autonomous replication sequence). It has high transformation efficiency (104 – 105 transformants/µg DNA) and high copy number (up to 100 copies/cell) but is also highly unstable (loss rate 10 – 20% per generation) due to lack of the 2µ origin. Thus, the plasmid can be easily lost during budding.
D. YCp (yeast chromosomal plasmid)
YCp is another extrachromosomal plasmid that replicates autonomously using a centromere region called CEN. It replicates as an additional chromosome and is thus maintained at a single copy. It has high transformation efficiency (104 – 105 transformants/µg DNA) but low copy number (1 copy/cell). It is stable and shows no segregation bias (loss rate 1% per generation). It is useful when studying effects of a single gene copy.
Compatibility with Bacterial Cells
Yeast plasmids that can also be maintained and propagated in bacterial cells are called ‘Shuttle vectors’. They contain a bacterial origin of replication (ori) and a bacterial selection marker (antibiotic resistance) in addition to yeast elements. Cloning, propagation, and plasmid preparation is much easier in bacteria than yeast due to the high copy number of plasmids and the fast growth and convenience of handling of bacterial cells. Hence, shuttle vectors are cloned, propagated, and stored in E. coli. When needed, shuttle vectors are isolated using plasmid preparation methods to be transformed into yeast cells for studies2.
Selection Markers in Yeast
Most yeast strains are insensitive to conventional antibiotics (as antibiotics are by nature ‘anti-bacterial’ agents) and hence do not function on antibiotic resistance selection. That being said, certain special antibiotics that affect eukaryotic cells (like G418 or Zeocin) have been used for yeast selection.
A more inherent capacity for auxotrophic selection is used more widely. Spontaneously occurring mutations in various metabolism genes have led to yeast cells incapable of producing certain nutrients, most commonly amino acids. Thus, these yeast cells rely on the particular nutrient provided in the culture medium. Yeast plasmids that contain the functional auxotrophic gene complement the yeast cells need for the nutrient when it is ‘dropped-out’ from the medium. This helps to select for yeast cells that have taken up the necessary plasmid4.
For example: if your yeast strain is a leucine mutant, it lacks a leucine metabolism gene and will not produce leucine inherently. Thus, you need to add a constant supply of leucine in the growth medium for growth of this strain. For cloning, use a plasmid that has a leucine metabolism gene on it so that successful transformation of the plasmid into your yeast strain can be monitored by ‘dropping-out’ leucine from the medium. Thus, the leucine-mutant parent yeast will not grow on the media plate unless it has the ‘leucine-supplementing’ plasmid in it. This helps for easy selection after transformation!
Points to consider:
- Only the yeast plasmids that have the complementary mutation in the selected yeast strain can be used, so select for your plasmids wisely.
- If the auxotrophic marker is produced in excessive amounts due to high copy number, it may cause metabolic burden on cell. In such cases, use a low copy number plasmid if compatible with your application. Additionally, some plasmids have been designed that contain partially defective promoters for auxotrophic markers that produce proteins in only small amounts, enough for selection but do not build up to excessive levels5.
- Also, some phenotypes may be masked or altered due to a selection marker. The only way to avoid this is to carefully choose the auxotroph based on available literature and known alternate functions of auxotrophic genes.
When Starting Recombinant Yeast Work:
- Determine auxotrophic mutations in your yeast strain
- Choose plasmids that have complementary auxotrophic markers
- Choose type of plasmid depending on required stability, copy number, and transformation efficiency parameters
- Clone your gene of interest into the plasmid and transform yeast cells using the Lithium acetate – Polyethylene glycol method
- Grow transformed cells on a drop out medium (all nutrients present minus the amino acid/nitrogen base present on the plasmid)
- Select for transformants and confirm presence of cloned gene
So, you can see that plasmids play a crucial role in not just bacterial but also yeast research. With this basic understanding of yeast plasmids, you can now easily choose the right plasmids for your application, translate molecular cloning and transformation strategies from your bacteria to your yeast work and work seamlessly from one model organism into another.
References and Further Reading:
- Gunge N.; Yeast DNA plasmids. Rev. Microbiol. 1983; 37:253-76
- Gnügge R., Rudolf F.; Saccharomyces cerevisiae Shuttle vectors. Yeast 2017; 34: 205–221.
- Volkert F., Broach J.; Site-Specific Recombination Promotes Plasmid Amplification in Yeast. Cell 1986, Vol. 46, 541-550.
- Pronk J. T. Auxotrophic Yeast Strains in Fundamental and Applied Research. Appl. Environ. Microbiol. May 2002; p. 2095–2100
- Erhart, E., and C. P. Hollenberg. The presence of a defective LEU2gene on 2µm DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. Bacteriol. 1983; 156:625–635.