Studying the Epigenome by Next Generation Sequencing
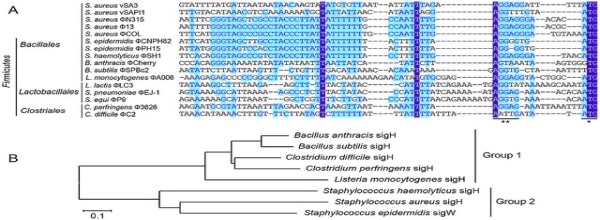
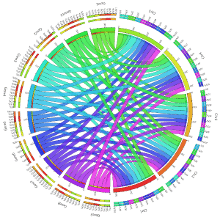
The epigenome has been in the research spotlight, and for good reason. Not only has it been associated with the developmental stages of an organism, but epigenetic alterations lead to disorders and have been linked to many human diseases. So, the question stands: what exactly is an epigenome? What Is the Epigenome? Simply put, the…