In this article, we share how CRISPR was first identified, the key discoveries that led to its development as a gene-editing tool, discover the astounding uses of this tool, and see how it has continued to be developed beyond gene-editing.
CRISPR-Cas9 in the Spotlight
CRISPR-Cas systems, such as CRISPR-Cas9, are adaptive immune response systems that protect prokaryotes from bacteriophages. They work by cleaving the nucleic acids of invading viruses, thus protecting prokaryotes from viral infections.
The use of CRISPR-Cas9 to edit genes was thrust into the spotlight in 2012 when George Church, Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang harnessed it as a tool to modify targeted regions of genomes. Given its potential to revolutionize gene editing, Science named CRISPR Breakthrough of the Year in 2015. In 2020, this technology earned Jennifer Doudna and Emmanuelle Charpentier the Nobel Prize in Chemistry.
But how was CRISPR first identified? When was its role in bacterial immunity discovered, and what other steps led to the development of this tool? Figure 1 shows an overview of some of the key events in the history of CRISPR, and we discuss these in more detail in the following sections.
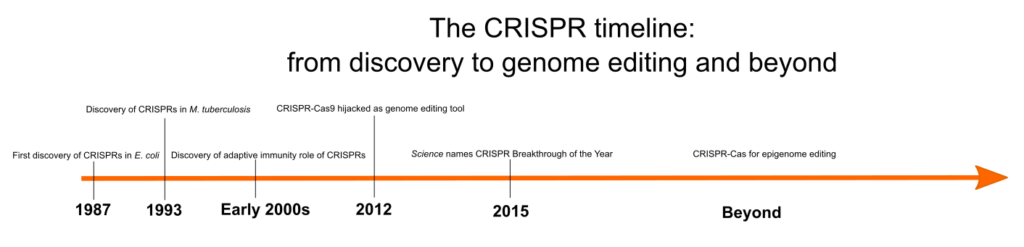
Discovery of CRISPRs
Clustered regularly interspaced short palindromic repeats, otherwise known as CRISPRs, are repeating DNA sequences in the genomes of prokaryotes, such as bacteria and archaea. CRISPRs were first identified in E. coli in 1987 by a Japanese scientist, Yoshizumi Ishino, and his team, who accidentally cloned an unusual series of repeated sequences interspersed with spacer sequences while analyzing a gene responsible for the conversion of alkaline phosphatase. [1] However, due to the lack of sufficient DNA sequence data, the function of these arrays remained a mystery.
In 1993, researchers led by J.D. van Embden in the Netherlands discovered that different strains of Mycobacterium tuberculosis had different spacer sequences between the DNA repeats. They characterized M. tuberculosis strains based on their spacer sequences, a technique known as ‘spacer oligonucleotide typing’ or spoligotyping. [2] Subsequently, these sequences were identified in several other bacterial and archaeal genomes. Researchers Francisco Mojica and Ruud Jansen were the first to refer to them as CRISPRs. [3]
CRISPR-Cas Systems as an Adaptive Immune Response
When CRISPR systems were first discovered, they were thought to be a novel DNA repair mechanism in thermophilic archaea and bacteria. [4] In the early 2000s, Mojica and coworkers noticed that the spacer sequences were similar to sequences found in bacteriophages, viruses, and plasmids. They discovered that viruses cannot infect bacteria possessing homologous spacer sequences, suggesting that these sequences play a role in the adaptive immune system in prokaryotes. [5]
In 2007, Rodolphe Barrangou, along with other researchers at Danisco, published a paper showing that new spacers derived from genomic sequences of phages were integrated into the virus-challenged bacteria. They also demonstrated that removing or adding particular spacers altered the phage resistance of the cell. [6]
How the CRISPR Adaptive Immune Response Works
When a virus infects a prokaryote, the spacer sequences in CRISPR arrays are transcribed to generate short CRISPR RNA (crRNA), which guides the CRISPR-associated sequence (Cas) protein to cleave complementary DNA or RNA viral sequences, depending on the type of CRISPR-Cas system. In this way, CRISPR-Cas systems function as a defense mechanism to prevent repeated infections by the same virus.
Other Components of CRISPR-Cas Systems
The role of Cas proteins as nucleases that cleave at specific sites was discovered by Makarova and colleagues, who conducted a comparative genomic analysis of CRISPR and Cas genes. They predicted that the function of CRISPR-Cas systems was similar to that of RNA interference, in which protein complexes silence genes by cleaving mRNA. [6] While some Cas proteins cleave DNA, others cleave RNA. For example, Cas9 cleaves DNA, while Cas13 cleaves RNA.
In addition to Cas proteins, protospacer adjacent motifs (PAMs) are essential components of CRISPR-Cas systems. As the name suggests, PAMs are short 2–6 base-pair sequences in the viral genome found adjacent to sequences targeted by Cas nucleases. Cas nucleases recognize PAMs and cannot cleave DNA unless a PAM is present. PAMs play an important role in ensuring that only foreign viral nucleic acids are cleaved but not CRISPR arrays. [8]
CRISPR-Cas9 Hijacked for Genome Editing
In 2012, George Church, Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang discovered that by designing guide RNA to target a specific region in the genome, “the CRISPR-Cas9 system can be used as a “cut-and-paste” tool to modify genomes. As a DNA-editing tool, CRISPR-Cas9 can remove or introduce new genes as well as silence or activate genes. CRISPR-Cas9 has been used to switch off genes that limit the production of lipids in microalgae, leading to increased lipid production and higher yields of biofuel. [9] In the near future, CRISPR-Cas9 may also be used to cure genetic disorders such as sickle-cell anemia and cystic fibrosis. [10] In fact, there is already a wide range of CRISPR applications in disease treatment, including cancer and infectious diseases.
A New Wave of CRISPR Tools
Since CRISPR’s debut as a genome editing tool, it has been adapted in various creative and beneficial ways. Point mutations can be used to inactivate one of the catalytic domains in Cas9, producing nickases, which cut just one strand of double-stranded DNA. Pairing two different nickases together can achieve the same double-strand break as fully active Cas9 but with enhanced specificity. [11]
Researchers have modified the Cas9 nuclease to perform targeted epigenome editing. This modified Cas9, known as enzymatically dead Cas9 (dCas9), can be linked to one of several enzymes that change the epigenome, e.g., DNA demethylases, methylases, or acetyltransferases. Like unmodified Cas9, dCas9 is directed to the targeted genomic region with a guide RNA. However, instead of cutting the DNA, the dCas9-enzyme complex modifies the epigenome at the site.
Epigenome editing can activate or repress transcription. Transcription can be activated by demethylating DNA using enzymes such as dCas9-Tet1 or by modifying histones using dCas9 linked to the histone acetyltransferase p300 enzyme. Conversely, transcription can be repressed by methylating DNA using DNA methyltransferase. Genes can also be silenced by linking dCas9 to enzymes that recruit corepressor proteins.
Similarly, gene expression can be activated (CRISPRa) or inhibited (CRISPRi) by fusing dCas9 to transcriptional effectors that activate or repress transcription.
Cas nucleases have also been engineered to decrease the level of off-target activity, generating high-fidelity Cas9 variants such as SpCas9-HF1 and eSpCas9-1.1.
Other CRISPR Nucleases
Cas9 isolated from Streptococcus pyogenes is the best-known and most well-used nuclease, but other nucleases have been identified that have different properties. Cas9 nucleases from other species have different PAM specificities and sizes, which can impact the ease of use.
Other flavors of Cas nucleases have also been found, including Cas13, which can be used to track, modify, and knockdown RNA in cells, and Cas12, which is more compact and better for editing AT-rich sites than traditional Cas9.
CRISPR in the Clinic
One of the obvious potentials of CRISPR is in the treatment of genetic diseases. This potential has been reached with the move of CRISPR from the research bench to the clinic as a therapeutic tool.
In 2019, CRISPR editing was trialed as a treatment for patients suffering from sickle cell disease and β-Thalassemia. [12] This success has led to many more patients receiving the same treatment.
And in 2023, CRISPR was used to modify CAR-T cells to treat teenagers with relapsed T-cell acute lymphoblastic leukemia. [13]
The Future of CRISPR-Cas9
CRISPR-Cas9 technology has its pros and cons. However, as researchers continue to use, refine, and expand the CRISPR toolkit, new applications and additional tools are constantly arising.
Whether you want to know how to get started with CRISPR or find new applications of this genome editing tool, head over to the Bitesize Bio CRISPR Research Hub.
CRISPR has revolutionized gene editing. And there’s a lot to learn. Get your free Gene Editing 101 eBook to get up to speed today.
Originally published June 2020. Reviewed and updated November 2023.
References
- Ishino, Y. et al. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. Journal of Bacteriology, 200, 7 (2018). e00580-17. doi: 10.1128/JB.00580-17.
- Sola, C. et al. High-throughput CRISPR typing of Mycobacterium tuberculosis complex and Salmonella enterica serotype Typhimurium. Methods in Molecular Biology, 1311 (2015). doi:10.1007/978-1-4939-2687-9_6.
- Morange, M. et al. What history tells us XXXVII. CRISPR-Cas: The discovery of an immune system in prokaryotes. Journal of Biosciences, 40 (2015). 221–223 https://doi.org/10.1007/s12038-015-9532-6.
- Makarova, KS. et al. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Research, 30, 2 (2002). 482–496. doi: 10.1093/nar/30.2.482.
- Mojica FJ, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005 Feb;60(2):174-82. doi: 10.1007/s00239-004-0046-3.
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007 Mar 23;315(5819):1709-12. doi: 10.1126/science.1138140.
- Makarova, KS. et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biology Direct, 1, 7 (2006). doi: 10.1186/1745-6150-1-7.
- Shah, SA. et al. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biology, 10, 5 (2013), 891–899. doi: 10.4161/rna.23764.
- Sharma, P.K. et al. Tailoring microalgae for efficient biofuel production. Frontiers in Marine Science, 5 (2018), 382. doi: 10.3389/fmars.2018.00382.
- Kotagama, O.W. et al. Era of genomic medicine: a narrative review on CRISPR technology as a potential therapeutic tool for human diseases. Biomed Research International, 2019 (2019). doi: 10.1155/2019/1369682.
- Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013 Sep 12;154(6):1380-9. doi: 10.1016/j.cell.2013.08.021.
- Frangoul H, et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N Engl J Med. 2021 Jan 21;384(3):252-260. doi: 10.1056/NEJMoa2031054. Epub 2020 Dec 5. PMID: 33283989.
- O’Leary K. Base-edited CAR T cells for pediatric leukemia. Nature Medicine. 2023 Jun 22. doi: 10.1038/d41591-023-00056-0.





