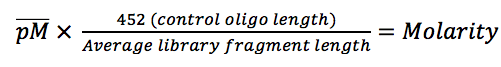There is no such thing as “junk” DNA
Until recently, vast areas of the genome had been denounced as “junk” DNA, because they do not encode proteins. However, it has become clear that these regions have a large diversity of other functions, from transcriptional and translational regulation to the protection of genes and genome integrity. The ENCODE project reported in 2012 that at least 78% of the genomic sequence (in humans) serve a specific function. Most of the functions are yet unknown, and there is strong interest in developing algorithms that help uncover the logical patterns within non-coding sequences. In this article, we’ll discuss a few different software options that you can use to identify conserved non-coding elements.
Non-coding sequence alignment using MULAN
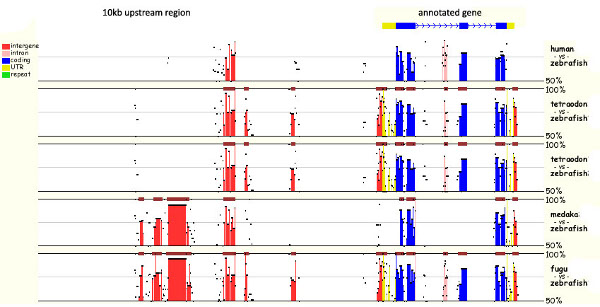
When aligning protein or mRNA sequences the software usually matches sequences by conservation, since these sequences are assumed to share common origin. However a characteristic of non-coding DNA is that functional elements can rearrange (change position, break up, invert) without losing their functionality, which makes them impossible to align with the same software. The free online software MULAN (MUltiple sequence Local AligNment and visualization tool) uses genes and surrounding regions to look for conservation in the non-coding DNA. The user has to provide sequence data from several species (depending on the depth of conservation you are looking for) for the same gene; for example, a gene plus 5kb of upstream sequence. Additionally, all exons in the area have to be annotated, because naturally they will show up as highly conserved areas. The alignment is performed pairwise, comparing each species with the species that was selected as a reference (see Figure 1: Screenshot of MULAN Output). The software can find patches of conservation that are in a different order or backwards, as is often the case with enhancer elements. Their position on the reference sequence is highlighted and the sequence alignment can be viewed and analyzed.
Uncovering synteny using Genomicus
Another way to approach conservation is taking synteny into account. Genes are said to be in synteny if the same genes occur in close proximity to one another across several species. A common feature of syntenic loci is that they also share regulatory elements (the most famous example of this is the hox gene clusters). An online genome browser that searches for syntenic genes is Genomicus. The user selects gene and species and the software calculates a phylogenetic tree based on this gene and shows the surrounding genes if they are in synteny. By ticking “CNE” (conserved non-coding elements) in the view menu, the software will also show areas of non-coding conservation between the syntenic genes.
Exploring synteny and conservation with UCNE base
Finally, conservation and synteny information can also be found conveniently presented in a browse-able database at UCNE base, “a database of ultraconserved non-coding elements and genomic regulatory blocks”. It includes many model species and a lot of data, so knowing exactly what you are looking for is mandatory in this case.
Any questions? Let us know in the comments section!
Check out our related article on identifying conserved elements in genes.
References:
Dimitrieva, S., & Bucher, P. (2012). UCNEbase–a database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic acids research, 1–9. doi:10.1093/nar/gks1092
Kikuta, H., Laplante, M., Navratilova, P., Komisarczuk, A. Z., Engström, P. G., Fredman, D., Akalin, A., et al. (2007). Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome research, 17(5), 545–55. doi:10.1101/gr.6086307