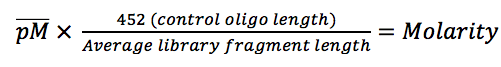Clustered Regularly Interspaced short palindromic repeats (CRISPR) associated proteins (Cas) are components of the prokaryotic adaptative immune system that protects them from viruses. Cas9 is the best known and widely used Cas protein for genome editing. However, the discovery of Cas13 proteins that bind and cut RNA opened the door to a new world of possibilities for genome editing. You can use Cas13 proteins in your research to knock down, modify or track RNAs in mammalian cells. Also, with the help of Cas13, you can detect specific RNAs in patient samples in a matter of minutes.
What is Cas13 and How Does it Work?
Discovery of Cas13 Proteins
CRISPR-Cas proteins are naturally found in bacteria and archaea. These proteins are classified based on their homology and function into Class 1 or Class 2. In 2015, new members of the class 2 proteins were identified. (1) One of the proteins identified was C2c2 (Class 2 candidate 2).
The C2c2 proteins differ from other Cas proteins by the presence of two Higher Eukaryotes and Prokaryotes Nucleotide-binding domains (HEPN) predicted to give the protein RNAse activity. Other groups have previously identified Repeat Associated Mysterious Proteins (RMAP) module (Cmr) Cas complex of proteins that target and cleave foreign RNA in Archaea. (2) But it was Abudayyeh and colleagues that took the task of characterizing the CRISPR-Cas protein C2c2/Cas13a. (3)
The Function of Cas13 Proteins
In bacteria, Cas13 proteins are a defense mechanism to silence foreign RNA. Unlike Cas9 or Cas12, Cas13 only binds and cuts ssRNA. Cas13 finds its target with the help of a guide-RNA (gRNA) otherwise known as a CRISPR-RNA (crRNA). The gRNA consists of a conserved sequence of approximately 70 nucleotides with a hairpin-like structure bound by Cas13, followed by a variable sequence of 29 nucleotides that is complementary to the target RNA.
Two HEPN domains provide the RNAse activity of Cas13. Mutations in these domains result in an inactive Cas13 protein, dead Cas13 (dCas13), that can bind but not cut RNA. Once Cas13 has cut its target RNA, it displays trans-collateral RNAse activity in bacteria. Even though the trans-nuclease activity can cut any RNA, independently of its sequence, Cas13 proteins from different bacteria species have a preference for specific sequences (figure 1).
Subclassifications of Cas13
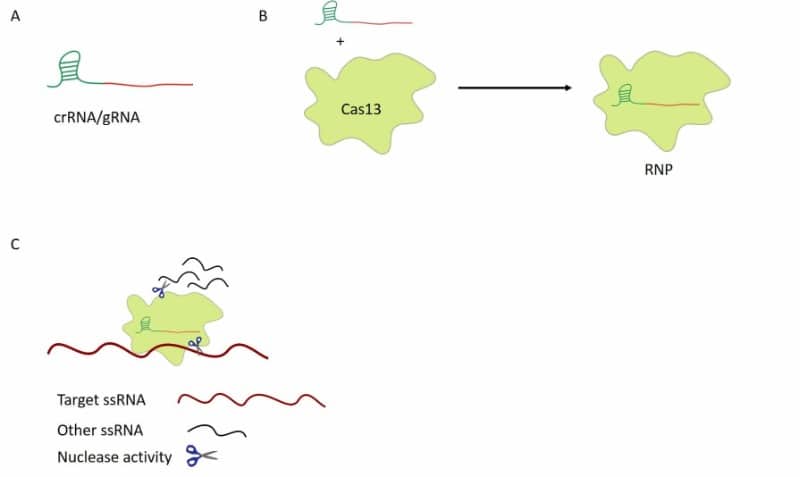
Cas13 proteins are present in at least 21 bacterial genomes (1). They all have two HEPN domains, but the size of the full-length protein can vary depending on which bacteria they come from. That is why they are subclassified as Cas13 a, b, c, d, etc. Also, because the sequence of the hairpin-like structure they bind to is different among the different species, the scientific name of the bacteria they come from is often included in the name; for example, LbaCa13a comes from Lachnospiracea bacterium. Figure 2 shows how different Cas13 proteins compare.
Since its discovery and characterization, this programmable RNA binding protein has changed the way we silence gene expression and how we study, modify, track, and detect RNA.
Applications of Cas13
Knocking Down RNA Using CRISPR-Cas13
The first application of Cas13 in eukaryotic cells was for knocking down (KD) mRNAs. But why use Cas13 to knock-down RNA?
- You can achieve more than 90% knock-down efficiency. This provides an excellent alternative for gene silencing when gene knock-out is lethal for the cell or when temporary gene repression is needed.
- You can get a better KD efficiency using Cas13 than RNA interference (RNAi). (4)
- There are fewer off-target effects with Cas13 than with RNAi. (4)
It was initially believed that the unspecific trans-cleavage activity of Cas13 was not present in eukaryotic cells. However, growing evidence suggests that the degree of off-target effects by Cas13 varies depending on which Cas13 protein is used. (5)
PspCas13b has fewer off-target effects than RfxCas13d, but also, the frequency of off-target effects varies according to the target RNA and the cell line. (5) To minimize off-target effects, it is important to carefully design gRNAs specific to the desired target RNA and always include negative controls in the experiments.
Tracking and Modifying RNA in Live Cells Using dCas13
Catalytically inactive Cas13 protein (dCas13) can bind RNA but cannot cleave it. This characteristic offers another way to use Cas13 to study and modify RNAs.
For example, you can increase the stability and translation of an RNA by fusing a methyladenosine transferase like TRMT6 to Cas13. (6) Or, if you fuse Cas13 to a fluorescent protein, such as GFP or mCherry, the location, and dynamics of RNA in the cells can be recorded by live-cell imaging. (7)
The greatest advantage of using dCas13 to study RNA is that the genome does not need to be edited to add other RNA sequences, like MS2, to track it, making dCas13 a quicker and easier system to use than the existing ones.
In the same way as when working with the active Cas13, it is important to demonstrate that the gRNAs used for dCas13 are specific. This can be done by using the dCas13 gRNA with the active form of the protein and seeing a reduction in the target RNA (measured by RT-qPCR).
Diagnosis and Treatment of Diseases
Due to its potential non-targeting effect, the trans-collateral activity of Cas13 does not make it the perfect tool for knocking-down RNA. However, it makes it perfect as a diagnostic tool, prompting the development of an in vitro diagnostic tool called Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK).
SHERLOCK allows the detection of specific RNA (viral or cellular) in a given sample. SHERLOCK can detect attomolar (10-18 moles per liter) amounts of RNA. Also, due to the specificity of Cas13, it is possible to detect single nucleotide mutations in the RNA. This allows the detection of RNAs carrying mutations and opens the possibility of using Cas13 to target RNAs associated with genetic diseases or cancer.
Where to Buy Cas13 Protein
Unfortunately, at the moment, there is only one commercial source of Cas13 proteins (McLab). Some companies can make and purify the protein in bacteria for you, but the cost can make it inaccessible for everyone. However, it is best to over-express the proteins from plasmids for research using mammalian cells. Loads of different plasmids are available from Addgene. Table 1 shows some of the most commonly used ones.
Plasmid name | Specie | Most common use | Source |
LwCas13a | Leptotrichia wadei | SHERLOCK | Addgene plasmid 91909 (for bacteria expression) |
PspdCas13b | Prevotella sp | Tracking RNA in live-cells | Addgene plasmid 132403 (for mammalian expression) |
RfxCas13d | Rumicoccus flavefaciens XPD3002 | Knocking down mRNA (warning, recent data suggest Cas13d has a very high non-targeting effect) | Addgene plasmid 109049 (for mammalian expression) |
Cas13 Summarized
From its discovery, Cas13 has become an important tool to help our understanding of the RNA world. Its small size, compared to Cas9, makes it an easy protein to work with. Its ability to bind and cut RNA makes it an excellent option for regulating gene expression when it is not possible to use Cas9.
dCas13 versions provide an easy way to modify specific RNAs. Even its trans nuclease activity can be wielded for RNA detection. I am sure this is just the beginning and that more applications of Cas13 will emerge in the future.
Do you use Cas13 in your research? Let us know how in the comments below.
References
- Shmakov S et al. (2015) Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 60(3):385–97
- Hale CR et al.(2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139(5):945–956
- Abudayyeh O et al. (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299): aaf5573.
- Granados-Riveron J et al. (2018) CRISPR-Cas13 Precision transcriptome engineering in cancer. Cancer Res. 78(15):4107–1413
- Ai Y et al. (2022) CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. NAR.; gkac159
- Tang T et al. (2021) Programmable system of Cas13-mediated RNA modification and its biological and biomedical applications. Front. Cell Dev. Biol. 9: 677587
- Yangs LZ et al. (2019) Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Molecular Cell 76: 981–997