T cells are central players in the adaptive immune response and have a key role in the defense against cancer. Some cancer cells manipulate the native immune checkpoint mechanism to evade detection by T cells. The programmed death-1 (PD-1) checkpoint receptor is one such example and is activated through the binding of the ligand PD-L1, which results in the inactivation of the T cell. Many cancers respond to the selective pressure exerted by the immune system by expressing the PD-L1 ligand.
Therapeutic antibodies have been developed by researchers to target and block the binding of PD-L1 to PD-1. Blocking the PD-1 receptor prevents the PD-L1-mediated T cell deactivation. This deactivation results in a boost of anti-tumor activity by T cells, resulting in a very clinically important target in oncology. While the use of antibodies can be an effective therapy, there are limitations.[1] Gene editing methods like CRISPR have the potential to overcome these limitations.
Ex-Vivo Editing of T Cells as a Therapeutic Strategy
The primary challenge to using therapeutic antibodies is the constant deactivation of the immune response by cancer cells during tumor progression. This requires the constant replenishment of antibodies through multiple rounds of treatment to achieve the desired therapeutic outcome.
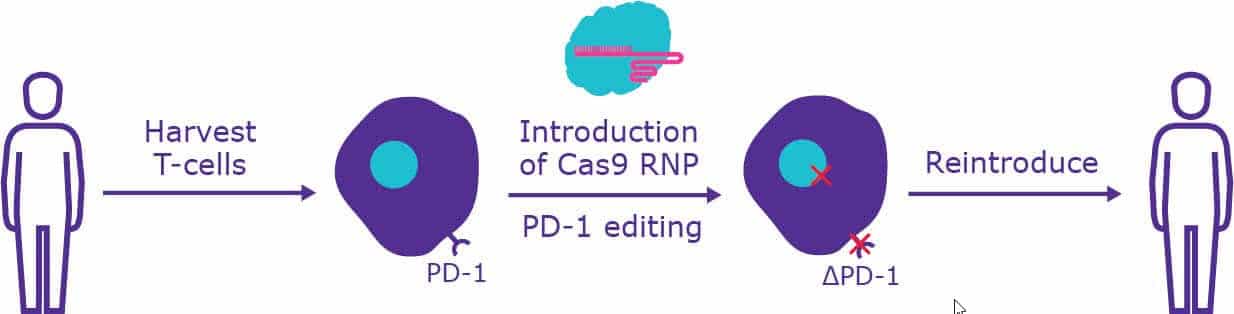
A more robust solution is to disrupt the PD-1 receptor through direct gene editing of a patient’s T cells. This involves the extraction of the T cells, knocking out the PD-1 receptor, and reintroducing the modified T cells into the bloodstream (Figure 1). Several gene-editing methods have been employed to achieve immune checkpoint inhibition including zinc finger nucleases (ZFNs), transcription-activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPRs). Each system has advantages and limitations.
CRISPR-based gene editing is safe, specific, easy to use and highly efficient, making it a particularly useful method for therapeutic applications. Studies are already showing the promise of this technique with CRISPR-mediated disruption of PD-1 enhancing the anti-tumor activity of cytotoxic T lymphocytes and CAR T cells. [2, 3]
It’s important to note that it is not necessary to isolate and edit every cell in the bloodstream of the patient for the therapy to be effective. Once activated, T cells proliferate, producing millions of cells that can then mount an immune response. Therefore, only a small fraction of the total T cell population of a patient needs to be altered.
Challenges of Genetically Reprogramming T Cells
CRISPR-based gene editing, while having advantages over other methods for editing primary cells, still presents some challenges. Here we review some of the major obstacles that include delivery methods, off-target effects, and editing efficiency.
Delivery Methods in T Cells
Application of CRISPR-mediated functional genomics in primary T cells has been limited largely due to inefficient vector delivery and the unwanted effects of delivery systems on cellular homeostasis. The delivery of the CRISPR-Cas9 machinery into cells has traditionally been performed using either viral vectors or plasmid-based delivery methods. Recent studies have shown that while CRISPR gene editing can be achieved in T cells using both of these methods, they are less efficient than in immortalized cells. [4-6] The advantages and challenges of each of these methods are discussed below.
Viral-Mediated Delivery
Lentiviral vectors permanently integrate into the host genome, which can result in off-target effects owing to the persistence of the CRISPR-Cas9 machinery in the host cell. The random nature in which the lentiviral vectors integrate into the host genome can also create undesirable and unpredictable side-effects.
Adenoviral (AdV) and adeno-associated viral (AAV) vectors can overcome the issue of random integration, as they do not integrate into the host genome. Instead, they exist as stable episomes within the host cell. However, these methods have their own limitations.
The small size of AdVs limits the amount of DNA that can be packaged within the virus to around 4.5 kb. [7] This can be problematic when using the relatively large nuclease SpCas9. This can be overcome by the use of an alternative Cas9 protein, SaCas9, which is around 70% smaller than SpCas9. [8]
A major drawback of AdVs is that they are known to trigger both innate and specific immune responses. While AAVs do not trigger the robust immune response as seen with AdVs, the capsid of AAVs can trigger a delayed immune response, which can cause CD8+ T-cell toxicity. [5]
In addition to the limitations for AAVs and AdVs listed above, viral vector development is expensive and time-consuming, making it difficult to implement in a large-scale therapeutic setting.
Non-Viral Delivery
Non-viral techniques to deliver plasmids in lymphocytes have previously yielded poor results. [9] However, the delivery of Cas9 protein as a ribonucleoprotein (RNP) complex using non-viral methods such as electroporation has shown to be successful in several studies [10, 11]. Delivery of large transgenes has been a particular problem in primary cells due to toxicity. A recent study has overcome this issue, showing Cas9 RNPs can be used to insert transgenes greater than one kilobase at specific loci in primary T-cell genomes with no harmful effects on cell survival and function. [12]
Delivery into primary T cells has also been improved by related advances such as the linking of CRISPR RNA (crRNA) and the trans-activating crRNA (tracrRNA) into a single guide RNA (sgRNA) that reduces the number of components that must be introduced into the target cell.
The delivery of RNP complexes has obvious advantages over DNA-based methods, as it does not rely on cellular expression machinery and is removed relatively quickly, limiting the potential for off-target effects. If selection markers are required to isolate edited cell populations (e.g. via antibiotic resistance or fluorescent tags) the use of an RNP complex can be more difficult. However, Cas9 protein that is fluorescently- labeled can now be purchased.
Reducing Off-Target Effects of CRISPR
Using Ribonucleoprotein Complexes to Minimize Off-Target Effects
The minimization of off-target effects is critical when cells are to be reintroduced into a patient. The use of an RNP complex can limit such unwanted effects. Unlike constructs delivered via lentivirus that integrate into the genome, the RNP complexes are present only transiently in the target cell. Rapid protein turnover in T cells degrades these Cas9 RNP complexes within 24 hours, [10] limiting the time Cas9 is active and reducing the opportunity for off-target activity.
Minimizing Off-Target Effects With Paired Nickases
Another method to limit off-target effects is the use of paired nickases. In this technique engineered versions of nucleases are used that can cut only one strand of DNA. For a double-stranded cut to occur with this system, each nickase must be paired with a complementary guide (gRNA) targeting the same region of DNA. This makes the probability of off-target cuts so low that they are effectively eliminated.
Benefits of Staggered Cleavage in Minimizing Off-Target Effects
Unlike other commonly used Cas9 nucleases, such as those isolated from Streptococcus pyogenes (SpCas9) and Staphylococcus aureus (SaCas9), the RuvC nuclease domain of Francisella novicida Cas9 (FnCas9) cleaves the target DNA in a staggered pattern, leaving four nucleotide overhangs at the 5’ end, and shows higher specificity. However, FnCas9 does not edit multiple targets in the human genome, having evolved to inactivate in certain mammalian chromatin contexts. The proximal CRISPR targeting method, also known as proxy-CRISPR, [13] exploits this curious characteristic of FnCas9. Proxy-CRISPR restores the nuclease function of FnCas9 in a target-specific manner allowing precise editing, even between identical sites in the same genome. The use of FnCas9, which is intrinsically more specific than SpCas9, further decreases the probability of off-target effects using the CRISPR system.
Improving Editing Efficiency in Primary Cells
Although the CRISPR-Cas9 system is remarkably efficient in immortalized cell lines, CRISPR-based genome editing has significantly reduced efficiency in T cells, even when the same gRNAs are used. [6] Current data suggest that the reduced efficiency is driven by a number of factors that include differences in rates of transfection, promoter compatibility, exonuclease activity, the production of interferons upon introduction of foreign genetic material and the high fidelity of DNA repair in human primary cells. Several modifications have been devised to improve editing efficiency with the CRISPR system in primary cells.
Choice of gRNAs and Editing Efficiency
In a two-part synthetic gRNA system, the universal tracrRNA and the specific crRNA remain separate. When these are combined into a sgRNA, editing efficiency has been shown to increase in primary human cells, including non-activated T cells. Further, recent studies have demonstrated that chemically altering sgRNAs increases their genomic editing efficacy in primary T cells, hematopoietic stem cells, and progenitor cells. [14]
Studies have shown that certain chemical modifications increase the stability of RNA. [15, 16, 17] Building on those data, sgRNAs have been modified with 2′-O-thionocarbamate, 2′-O-methyl, 2′-O-methyl 3’phosphorothioate, or 2′-O-methyl 3′ thioPACE at multiple nucleotides at both ends. When introduced together with Cas9 (in the form of mRNA or RNP), modified sgRNAs offer a distinct advantage for gene editing in human cell lines and primary unstimulated T cells. It is worth noting that the efficacy of the specific chemical modification is dependent on the sequence, cell type, and delivery conditions.
RNPs and Editing Efficiency
In addition to improved delivery, Cas9 RNP complexes exhibit high editing efficiency in T cells. [10] Transfection of these complexes using nucleofectin obtained a target knockout rate of 90% [18], which can be further enhanced using electroporation. The modified electroporation approach does not require T-cell activation before transfection and therefore can be utilized in studies of T-cell differentiation while maintaining cell viability.
Combining RNPs and Paired Nickases to Improve Editing Efficiency
As discussed above, the use of paired nickases can reduce off-target effects. When delivered as a paired-nickase RNP complex, this method has the added benefit of high editing efficiency.
In collaboration with MaxCyte, we showed that the paired-nickase system leads to >80% reduction in PD-1 protein expression over 5 days in peripheral blood mononuclear cells (PBMCs). The paired-nickase system spreads out its indels over a wider area than the single spCas9 nuclease system, leading to greater phenotypic efficacy. Other clinically relevant targets in CD8+ T cells, such as CTLA4, TIM-3, and TRAC, have also been successfully edited with high efficiency and specificity using the paired-nickase RNP system.
Conclusion
Gene editing of T cells is a powerful new, effective tool for immuno-oncology therapy. As with any new technology, challenges exist when implementing this tool, including the delivery of the CRISPR machinery, the potential for off-target effects, and low editing efficiency. However, these issues can be overcome through the careful selection of the CRISPR machinery and mode of delivery.
More CRISPR Resources
CRISPR Articles
- CRISPR Gene-editing: Considerations and Getting Started
- How to Validate a CRISPR Experiment
- How to Understand CRISPR Formats and Their Applications
CRISPR Webinars
- Engineering Vero Cell Lines Using CRISPR to Increase Production of Viral Vaccines
- Efficient Generation of Gene-edited Mouse Models and Cell Lines Using Synthetic sgRNA
- Optimizing CRISPR mouse model pipelines – modified synthetic sgRNAs
- Efficient Generation of Gene-edited Mouse Models and Cell Lines Using Synthetic sgRNA
CRISPR eBook
CRISPR Tools
And you should also consider the following Sigma-Aldrich® tools:
- CRISPR Design Tools
- CRISPR Controls
- CRISPR Essentials
- Primary T cell Editing Using Nickase RNPs
- Advanced Genomics Resource Center
- Sigma-Aldrich® Advanced Genomics
- Additional CRISPR Webinars
Discover more about CRISPR in the Bitesize Bio CRISPR Research Hub.
References
- Michot, J.M., et al. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur J Cancer. 54:139-148 (2016). DOI: 10.1016/j.ejca.2015.11.016.
- Zhao, Z. et al. CRISPR knock out of programmed cell death protein 1 enhances anti-tumor activity of cytotoxic T lymphocytes. Oncotarget. 9(4):5208-5215 (2017). DOI: 10.18632/oncotarget.23730.
- Choi, B.D., et al. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J. Immunotherapy Cancer 7, 304 (2019). doi:10.1186/s40425-019-0806-7
- Li, C. et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol 96, 2381-93 (2015). DOI: 10.1099/vir.0.000139
- Samulski, RJ. et al. AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol 1:427-51 (2014). DOI: 10.1146/annurev-virology-031413-085355
- Mandal, P.K. et al. Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell 15(5):643-52 (2014). DOI: 10.1016/j.stem.2014.10.004.
- Wu, Z., et al. Effect of genome size on AAV vector packaging. Mol Ther 18:80–6 (2010). DOI: 10.1038/mt.2009.255
- Ran, F.A., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–91 (2015). DOI: 10.1038/nature14299
- Su, S. et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep 6, 20070 (2016). DOI: 10.1038/srep20070.
- Kim, S., et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24, 1012-9 (2014). DOI: 10.1101/gr.171322.113.
- Schumann, K. et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. PNAS 112(33):10437-42 (2015). DOI: 10.1073/pnas.1512503112.
- Roth, T. L. et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405-409, (2018). DOI: 10.1038/s41586-018-0326-5.
- Chen, F. et al. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun 8, 14958 (2017). DOI: 10.1038/ncomms14958.
- Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985-9 (2015). DOI: 10.1038/nbt.3290.
- Deleavey GF, and Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol 19:937–954 (2012). DOI: 10.1016/j.chembiol.2012.07.011
- Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther 24:374–387 (2014). DOI: 10.1089/nat.2014.0506
- Dellinger, D.J., et al. Solid-phase chemical synthesis of phosphonoacetate and thiophosphonoacetate oligodeoxynucleotides. J. Am. Chem. Soc. 125:940–950 (2003). DOI: 10.1021/ja027983f
- Oh, S. A., et al. Ribonucleoprotein Transfection for CRISPR/Cas9-Mediated Gene Knockout in Primary T Cells. Curr Protoc Immunol 124, e69 (2019). DOI: 10.1002/cpim.69