If you’re doing research in biotechnology or synthetic biology, it won’t take long until you encounter the pBAD or ParaBAD promoter in one of your experiments. During my Master’s, it quickly became a staple, showcasing its importance and prevalence in controlled gene expression systems.
However, it also didn’t take long to understand its limitations and easy-to-mess-up nature.
This article explains how to express proteins using the pBAD promoter and how to avoid common issues such as arabinose catabolism, glucose repression, and leaky expression.
Why Do We Use Inducible Promoters?
Production of recombinant proteins is a common practice in most bioscience research. Common protein targets are insulin, antibodies, fluorescent proteins, or any other protein of interest.
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
However, such protein expression experiments wouldn’t have been possible without inducible promoters. [1]
The majority of heterologously expressed proteins are toxic to the host bacteria, even when it’s a native gene that’s being overexpressed.
Because expression is burdening the cell, it could:
- Inhibit cell growth
- Causes cell death
- Lead to the loss of the plasmid
Inducible promoters help solve these problems.
They enabled us to control when and at what rate the protein of interest is expressed.
Contrary to constitutive promoters that are always active, inducible promoters could be switched on and off under specific conditions (ligand concentration, pH, temperature, light, etc). [1]
Such promoters made biology much cooler and easier to engineer. One example is a biosensor for water contaminants [2] that can detect harmful chemicals in real time and light up with a fluorescent glow, like a tiny high-tech firefly signaling toxins. Check out this video of how the biosensor works.
How the pBAD Promoter Works
PBAD is a medium-strength inducible promoter. [3,10] Promoter strength reflects the expression level of a promoter, which is influenced by RNA Polymerase binding affinity, transcription factor interactions, and sequence architecture. You can think of pBAD in the middle of the promoter spectrum between the T7 (strong) promoter and the HexR/Pzwf1 (weak) promoter.
Expression under pBAD is positively and negatively regulated by the AraC protein. [4] That means AraC is wearing two hats in this process. It can either repress or activate genes downstream of pBAD.
It depends on the presence or absence of l-arabinose (the inducer) in the culture media.
Its mechanism of action is a bit similar to that of the infamous lac operon.
When arabinose binds to the AraC, it leads to a conformational change that opens the DNA loop, allowing RNA polymerase to bind and start transcription. [4,5]
Without induction (no l-arabinose added), AraC binds to DNA as a repressor, prohibiting transcription initiation. Figure 1 below shows a schematic overview of the pBAD expression system.
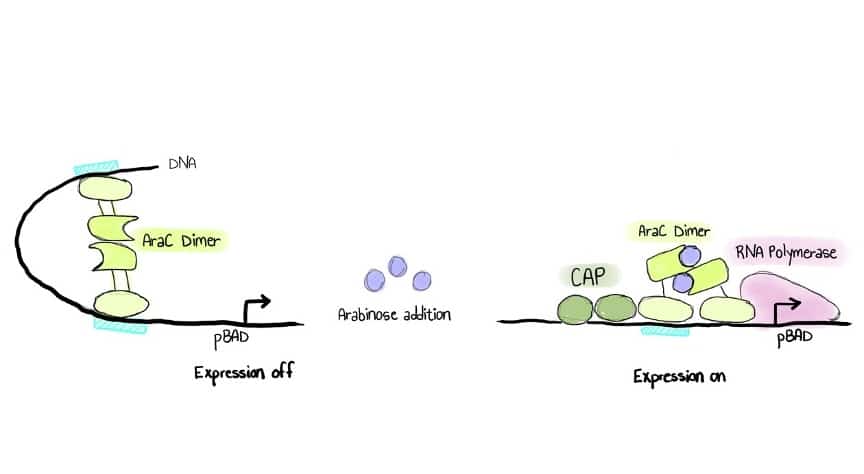
Figure 1. A simple schematic of the pBAD expression system. (Image credit: Mariam Ezzelarab.)
pBAD is a tightly controlled promoter that is useful in many experiments, [10,11] such as:
- Expressing highly toxic genes
- Studying null mutations of essential genes
- Optimizing protein solubility in E. coli
- Expressing membrane proteins
- Gene function analysis
- Synthetic biology applications
However, it can be tricky to adopt if you have never used it before, and there are some common issues to be aware of. Here are some ways in which pBAD can spoil your experiments and how to avoid them.
Common Issues with pBAD and How to Avoid Them
Leaky Expression
Expression leakiness occurs when an inducible promoter drives gene expression even when the inducer is absent.
Leakiness may not pose a big issue in some experiments, but in others, it can be a game-changer, either way, its impact is worth considering.
One of my first times working with pBAD, I was building a toggle switch in E. coli.
I was expecting the expression of red fluorescent protein in case of induction (l-arabinose added to media) and green fluorescent protein, if the inducer was not added.
However, the results weren’t what I anticipated.
The pBAD promoter’s leaky expression was evident in my samples, with red fluorescent protein appearing even when the arabinose was not added to the media, as you can see in Figure 2 below. I was stunned. My professors and supervisors? Not so much.

Figure 2. Quantitative comparison of fluorescent protein induction with and without l-arabinose. There is fluorescence corresponding to the expression of red fluorescent protein (the red line) despite no arabinose being present. (Image credit: Mariam Ezzelarab.)
They knew the truth: biology isn’t binary. The cellular environment is chaotic, and this is a clear example of this stochasticity. They’d seen it all before, especially with the pBAD promoter.
Arabinose Catabolism
L-arabinose is a sugar. It’s not exactly a bacterial gourmet dish (like glucose), but they’ll still chow down on it when it’s on the menu!
This results in a problem.
E. coli can consume the inducer [3,5,6], causing inconsistent expression due to the depletion of arabinose in the medium over time.
To prevent this issue, always avoid using wild-type E. coli with pBAD. Instead, opt for host strains mutated to be deficient in L-arabinose catabolism, such as TOP10 or LMG194. [3]
Glucose Repression
When working with pBAD systems, glucose is both a friend and an enemy. [7]
As mentioned above, bacteria favor glucose much more than any other source of carbon.
We can make good use of this feature by adding glucose in the media to further repress the gene expression in the absence of the inducer [3,10] and act as an additional barrier that prevents gene expression during the off state.
However, glucose could be a problem in the on state, simply because the bacteria will choose to uptake glucose from the media instead of l-arabinose.
So, the inducer will not go inside the cell and will not induce gene expression. Figure 1 in the iGEM registry for pBAD illustrates this well.
This issue is easy to solve.
All you need is to be cautious when selecting a bacterial medium for your experiment. Don’t just consider the strain’s optimal media for growth, but also check the ideal media composition for the optimal promoter induction.
In one of my experiments, I expressed beta-carotene under the control of the pBAD promoter.
While temperature, induction time, and inducer concentration were the exact same, different media compositions were tested. Check out Figure 3 below to see how the protein of interest concentration varied from one media to another due to different compositions.

Figure 3. Comparative analysis of beta-carotene production in different bacterial media. (Image credit: Mariam Ezzelarab.)
For my experiment, LB Broth (Luria/Miller), LB Broth (Lennox), and Terrific Broth resulted in the maximum yield as they contained no glucose to inhibit pBAD induction.
All-or-None
pBAD gives rise to a phenomenon known as “all-or-none”.
This means at sub-saturated inducer concentrations (where the cell hasn’t reached full saturation and still has room for more), instead of getting a uniform and incremental level of induction, you end up with a mix.
Some of the cells are fully induced while others go uninduced. As you can see in Figure 4, when protein expression is measured at the level of the population it gives the appearance of a smooth gradient of expression. However, that doesn’t mean individual cells are expressing more protein, but more cells just start to be induced.
This could be critical in some experiments because of inconsistent protein production, making it challenging to study dose-response relationships and leading to misinterpretation of the results.
There is no straightforward mitigation for this phenomenon. However, here are some tips that might help you design a more efficient and precise system.
A handful of iGEM competition teams concluded that the pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high-copy vector, but allows for gradual induction when cloned into a low-copy vector. [8,9]
Another method involves increasing the expression of the arabinose transporter. [12] This should reduce the threshold concentration required for induction and ensure uniform uptake across the population and thus reduce the heterogeneity of the cell population.

Figure 4. All-or-none gene induction of pBAD. A simple drawing showing all-or-none behavior. At the level of single cells, increasing the inducer changes the ratio of ON and OFF cells. (Adapted from Carla J. Davidson et al. (2008); image credit: Mariam Ezzelarab.)
Using pBAD In Summary
pBAD could express some targets fantastically and some dreadfully, proving efficient in some studies and disastrous in others. This is probably true for all promoters; you’ll never know until you try it out.
A perfect inducible promoter would use a cheap inducer, be harmless to the host’s metabolism, require no special transporters, and have a high dynamic range, low leakiness, fast kinetics, high specificity, and tunable expression levels. Even better, it would work across many species and scale up easily for industrial use.
Of course, a system this perfect probably doesn’t exist!
But if you’re wondering what promoter you should try next for your protein expression, you now have a rough idea of whether pBAD might work and how to troubleshoot some issues that might occur as you optimize your protein expression
References
- Gearing M. Plasmids 101: Inducible Promoters. Addgene. Published 18 January 2018. (Accessed 4 February 2025)
- Jung JK et al. (2020). Cell-free biosensors for rapid detection of water contaminants. Nature Biotechnology. 38:1451–1459
- pBAD Bacterial Recombinant Protein Vector. Vector Builder. (Accessed 4 February 2025)
- Schleif R. (2003). AraC protein: a love–hate relationship. BioEssays 25:274–282
- Schleif R. (2021). A Career’s Work, the l-Arabinose Operon: How It Functions and How We Learned It. EcoSal Plus 10
- Miyada CG et al. (1984). Regulation of the araC gene of Escherichia coli: catabolite repression, autoregulation, and effect on araBAD expression. PNAS 81:4120–4124
- Simcikova M et al. (2014). On the dual effect of glucose during production of pBAD/AraC-based minicircles. Vaccine. 24:2843–2846
- CU Boulder iGEM Team 2015. Testing response of pBAD promoter with Arabinose. iGEM. (Accessed 4 February 2025)
- iGEM Teams. Inducible pBad/araC promoter. Registry of Standard Biological Parts. (Accessed 4 February 2025)
- Guzman LM et al. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology 177:4121–4130
- Brautaset T et al. (2008). Positively regulated bacterial expression systems. Microbial Biotechnology 2:15–30
- Széliová D et al. (2016). Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains. Journal of Biotechnology 236:1–9
- Davidson CJ et al. (2008). Individuality in Bacteria. Annual Review of Genetics 42:253–268
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.