We’re all familiar with DNA and RNA, the two naturally occurring nucleic acids that store genetic information. But have you heard of xeno nucleic acids (XNAs)? If you’re unfamiliar with these synthetic nucleic acid analogs, don’t panic!
We’re going to take you through what XNAs are, how they can be applied in biological research, and how you can start using them in your experiments.
XNAs can be used in a host of genetic engineering applications. These include expanding the genetic code and creating aptamers—synthetic oligonucleotides or peptides that specifically bind a target molecule such as an enzyme. XNAs are also more stable than their natural counterparts (read on to find out why).
XNAs have also been used as tools for molecular diagnostics. They are becoming increasingly popular for developing novel diagnostic assays by incorporating synthetic biology approaches for detecting target nucleic acid sequences.
What Are Xeno Nucleic Acids (XNAs)?
Although XNAs are not found in nature, they resemble naturally occurring nucleotides, with some notable differences.
Nucleotides comprise three main parts:
- the phosphate group;
- the furanose sugar; and
- the nitrogenous base.
All three parts can be synthetically modified or engineered by incorporating different groups that don’t occur naturally.
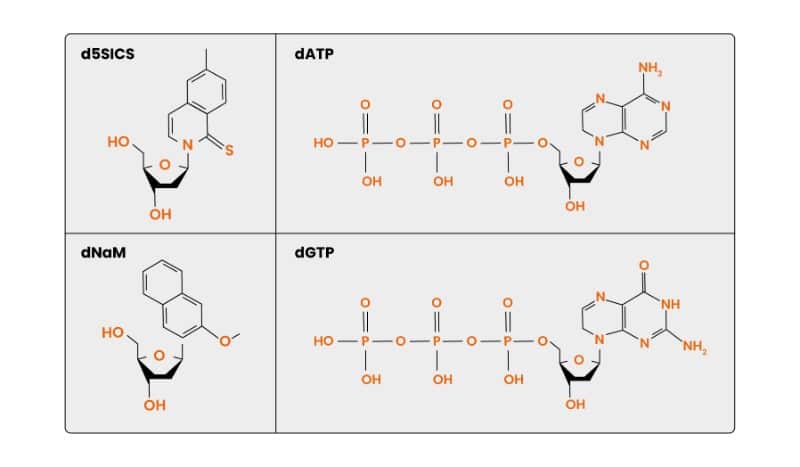
Figure 1 shows two widely-used XNAs: d5SICS and dNaM, and their naturally occurring counterparts, dATP and dGTP.
So, XNAs are non-standard DNA and RNA analogs with altered sugar rings, bases, or backbones that obey the Watson–Crick pairing rules and allow unnatural base pairing. What’s more, they’re still able to store and pass on genetic information.
How Are XNAs Used?
Owing to their variety, XNAs offer many possibilities for novel applications in synthetic biology, therapeutics, molecular diagnostics, and biotechnology. We’ve touched on some of these applications below.
1. Expanding the Utility of Nucleic Acids with XNAs
With advances in nucleic acid chemistry, XNAs have been developed with improved base-pairing stability compared with natural nucleic acids and increased activity in living tissues, thereby expanding their utility in the lab, and leading to the development of several nucleic acid therapeutics. Here are a couple of examples of how this has been achieved.
Altering the Phosphodiester Backbone to Increase Stability
The phosphodiester backbone of DNA can be modified by substituting a boron or sulfur atom in place of the oxygen atom that is single bonded to the phosphorus atom. These yield boranophosphate [1] and phosphorothioate [2], respectively, and polymers composed of these XNAs are resistant to nucleases, thus increasing their stability for therapeutic applications. [3]
Substituting the Furanose Ring to Increase the Melting Temperature
Another application involves substituting the furanose ring (ribose or deoxyribose) with a different sugar moiety.
The locked nucleic acid (LNA) is a good example of this where a covalent linkage of a methylene group to the 4’ carbon, and the oxygen of the 2’ carbon (an extra bridge connecting them) “locks” the five-carbon sugar ring into confirmation stabilizing LNA–DNA and LNA–RNA duplexes.
This increases the melting temperature compared with DNA–DNA and DNA–RNA duplexes. [4]
Incorporating LNAs into oligonucleotide primers or probes can shorten them but still allow for annealing at higher temperatures. You would need a longer oligo if you were using natural nucleotides.
As a consequence, PCR primers incorporating LNAs can enhance PCR amplification sensitivity under a wide range of conditions—LNAs have been shown to enhance real-time PCR and DNA sequencing. [4]
2. Expanding the Genetic Alphabet
Perhaps the most straightforward application of XNAs is their utility in creating an expanded genetic code, which is of great interest in synthetic biology. With six nucleotides instead of four, you can theoretically form 216 possible codons. This enables greater information storage in DNA, and, thereby, the potential for the expanded genetic code to produce novel biochemical pathways that are constrained in nature.
How is this achieved? Modifying the nucleobase to maintain Watson–Crick hydrogen bonding using thymine or cytosine analogs is one strategy. Another is to use hydrophobic bases that “pair” with each other and simulate the shape of paired natural bases. [3]
Polymerase Synthesis with XNAs
The first step in expanding the genetic code is to get existing enzymes to incorporate XNAs into replicating DNA or synthesized mRNA. [5] The pair formed between the XNA known as d5SICS and another XNA called dNaM, or between d5SICS and an XNA known as dMMO2, can be incorporated into DNA polymers in vitro [6] and transcribed in vitro using the T7 RNA polymerase. [7]
The next step is to get a living cell to incorporate XNAs into DNA, tRNA, and mRNA and produce an unnatural base pair within a codon that, in turn, codes for an unnatural amino acid (UAA). One study, for example, used engineered E. coli to incorporate base pairs of dNaM and another XNA called dTPT3 (a d5SICS analog) into replicating DNA. These XNAs were also transcribed into tRNA and mRNA, and with cognate amino acyl transferases, incorporated the UAA pyrrolysine into a modified superfolder GFP. [8]
3. Aptamer-based Therapeutics
XNAs also have useful biotechnology applications, as they can be used to create novel aptamers that are more stable than their natural counterparts. The in vitro screening method known as Systematic Evolution of Ligands by Exponential Enrichment (SELEX) can be used to create novel, high-affinity ligands from oligonucleotides. [9]
These methods have been exploited for therapeutic activity, but oligonucleotides are rapidly degraded when introduced into the bloodstream, which doesn’t make for a very good therapy! You can, however, conduct SELEX using XNAs, and the ligands incorporating these will be much more stable.
In one study, an aptamer using XNAs containing hexitol instead of deoxyribose was found to be DNase I-resistant in human serum and to bind a protein known as vascular endothelial growth factor (VEGF). [10] VEGF is an interesting target considering its role in tumor growth and inflammatory diseases, and this XNA-based aptamer has the potential for therapeutic use.
4. Innovations in Molecular Diagnostics
XNAs also have utility as molecular diagnostic tools. As mentioned earlier, LNAs have higher hybridization melting temperatures than their natural nucleotide counterparts, enabling shorter PCR primers or PCR probes to be made. This allows one to achieve sufficient sensitivity and specificity by amplifying shorter stretches of DNA.
A good example of a molecular diagnostics innovation is an assay developed to detect mutations in the BCL1 and BCL2 genes in lymphomas. [11] The shorter amplicon (that’s the stretch of amplified DNA) was a solution to previous limitations using primers and probes composed of natural nucleic acids. In previous work, longer amplicons were needed to achieve the specificity required for a clinical-grade assay.
LNAs enable shorter primers and probes and, hence, shorter amplicons. This approach is especially helpful if your PCR is targeting a GC-rich region of DNA. [12]
Similarly, the XNAs isoguanosine (iG) and isocytidine (iC) pair with a higher affinity than natural guanosine and cytidine. [13] This has the effect of stabilizing the duplexes in which they are incorporated. Luminex’s MultiCode®-RTx Assay Technology [14] uses iG and iC to enable probe-free real-time PCRs, owing to the specific hybridization of iC and iG to each other and not to their naturally occurring counterparts.
How to Make Synthetic DNA
Incorporation of XNAs into DNA and RNA in vitro and in vivo is achievable but requires the engineering of a suitable cellular host. [8]
So how do you get started?
If you’re OK with an in vitro approach, then you’re in luck. One relatively simple approach is to have DNA or RNA polymers chemically synthesized to incorporate XNAs.
QIAGEN will do this for you. Their LNA Oligo Optimizer tool allows you to enter your DNA sequence with LNAs into a web interface and determine the melting temperature; you’ll also be able to see if there will be any secondary structure or self-complementation. XNAs, such as iG and iC, can be incorporated into PCR primers and probes by IDT (Integrated DNA Technologies). [15]
Another approach is to generate DNA or RNA fragments in vitro using PCR or in vitro transcription. The hydrogen bond-forming XNAs triaza-isoG (dP) and nitropyridone (dZ) can be incorporated into amplified DNA using Taq polymerase. Likewise, the hydrophobic nucleobase XNAs dNaM and d5SICS are also processed by Taq polymerase. [16]
dNaM and d5SICS can be transcribed in vitro by T7 RNA polymerase. [7] To create DNA or RNA fragments incorporating these, you simply add the XNA to your conventional PCR mix and let the enzymes do the work!
Where Next?
The in vivo approach for using XNAs is relatively new, and synthetic biology is a rapidly advancing field. Synthetic biology can create synthetic lifeforms using a parallel biology composed of XNAs, and enhance existing lifeforms by adding XNAs to the genetic code.
There is proof of concept insofar as we can create novel organisms with expanded genetic alphabets, but how that affects metabolism, biochemistry, and the subcellular organization of synthetic life is still to be determined. [17,18]
Please feel free to enter any tips or tricks you’ve discovered for creating XNA polymers in the comments field below; we’d love to hear from you!
References
- Micklefield J. et al. (2001). Backbone modification of nucleic acids: synthesis, structure and therapeutic applications. Curr Med Chem. 8:1157-79.
- Wilson C and Keefe AD. (2006). Building oligonucleotide therapeutics using non-natural chemistries. Curr Opin Chem Biol. 10:607-14.
- Appella DH. (2009). Non-natural nucleic acids for synthetic biology. Curr Opin Chem Biol. 13:687-96.
- Petersen M and Wengel J. (2003). LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 21:74-81.
- K. Dhami, et al. (2014) Systematic exploration of a class of hydrophobic unnatural base pairs yields multiple new candidates for the expansion of the genetic alphabet. Nucleic Acids Res. 42:10235-10244.
- Malyshev DA, et al. (2012) Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet. Proc Natl Acad Sci U S A. 109:12005-10.
- Seo YJ, et al. (2009) Matsuda S, Romesberg FE. Transcription of an expanded genetic alphabet. J Am Chem Soc. 131(14):5046-7.
- Zhang Y, et al. (2017). A semi-synthetic organism that stores and retrieves increased genetic information. Nature. 551:644-647.
- Chai C, et al. (2011). SELEX (Systematic Evolution of Ligands by EXponential Enrichment), as a powerful tool for deciphering the protein-DNA interaction space. Methods Mol Biol. 754:249-258.
- Eremeeva E, et al. (2019). Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res.. 47: 4927–4939
- Sørensen CD, et al. (2014). Common consensus LNA probe for quantitative PCR assays in cancer: Vehicles for minimal residual disease detection in t(11;14) and t(14;18) positive malignant lymphomas. J. Immunol. Meth., 406:131-136.
- Wang Q, et al. (2013). Molecular beacons of xeno-nucleic acid for detecting nucleic acid. Theranostics. 3:395-408.
- Chen X, et al. (2001). Stability and Structure of RNA Duplexes Containing Isoguanosine and Isocytidine. J. Am. Chem. Soc. 123:1267-1274.
- MultiCode®-RTx Assay Technology | Luminex Corporation, accessed 8-22-2021
- Integrated DNA Technologies – Modified Bases modifications (idtdna.com), accessed 8-22-2021
- Marx A and Betz K. (2020) The Structural Basis for Processing of Unnatural Base Pairs by DNA Polymerases. Chemistry. 26:3446-3463.
- Nie P, et al. (2020). Synthetic Life with Alternative Nucleic Acids as Genetic Materials. Molecules. 25:3483.
- Budisa N, et al. (2020). Xenobiology: A Journey towards Parallel Life Forms. ChemBioChem. 21:1439-4227.