Whereas DNA can survive for millennia, RNA is short-lived, which can make total RNA extraction tricky, because RNA is prone to degradation by enzymes called RNases, which are everywhere.
Therefore, isolation of total RNA from cells and tissues requires a method that will efficiently isolate the RNA from the samples while also minimizing RNA degradation.
What is total RNA?
Total RNA, as you might expect, is all the RNA molecules found inside a cell. This includes:
Messenger RNA (mRNA): long protein-coding messenger RNA transcripts, which serve as the instantaneous readout of cellular gene expression under particular conditions
MicroRNA (miRNA): a myriad of other smaller noncoding RNA molecules, many of which are involved in regulating and silencing gene expression.
Ribosomal RNA (rRNA): a key component of ribosomes and critical for protein synthesis.
Transfer RNA (tRNA): another critical component for protein synthesis. These RNA molecules transport amino acids to the ribosome and base pair with the mRNA to ensure the correct amino acid is added to the protein being synthesized.
What Are RNases?
Ribonucleases (RNases) are enzymes that degrade RNA. These enzymes are very problematic when isolating or working with RNA because RNases are ubiquitous (found everywhere), difficult to eliminate, and very destructive. Therefore you need to ensure you are working RNA-free when extracting total RNA.
Isolating total RNA, minus the RNases using TRIzol®
Fortunately, there are ways to inactivate RNases and prevent your precious RNA samples from being destroyed. One method to extract RNA while also inhibiting RNases is the Guanidinium thiocyanate-phenol-chloroform extraction method first applied for RNA extraction by Piotr Chomczynski and Nicoletta Sacchi.
TRIzol® is a monophasic mixture of phenol and guanidine isothiocyanate commonly used for RNA extraction and is a powerful protein denaturant that breaks down protein cell components and inactivates all enzymes, including RNases.
TRIzol extraction typically uses acidic phenol–chloroform to confine total RNA in a clear aqueous phase while proteins and cell debris end up in the pink organic layer. RNA can be recovered by precipitation with isopropanol, washed, and then redissolved in water. TRIzol is a brand name, and many other suppliers supply their own version including:
- RNAzol®
- QIAzol
- TriPure™
It is also possible to make your own phenol and guanidine isothiocyanate mixture.
Protocol for total RNA extraction from Cells and Tissues Using TRIzol
1. Cell Lysis
If you are isolating RNA from tissues, you will need to homogenize the sample first in 1 ml of TRIzol reagent per 50 to 100 mg of tissue using a homogenizer. The sample volume should not exceed 10% of the TRIzol volume.
If you are isolating RNA from adherent cells grown in culture, rinse the cells with ice-cold PBS and lyze cells directly in a culture dish or flask by adding 1 ml of TRIzol reagent per 10 cm² area and scraping with cell scraper or pipette tip.
Pass the cell lysate several times through a pipette and vortex thoroughly. If your culture is composed of suspension cells, spin cells down to remove old media, wash in PBS lyse cells with 1 ml TRIZOL for up to 10 million cells by pipetting up and down several times.
2. Incubation and Phenol–Chloroform Separation
Incubate the homogenized sample for 5 minutes at room temperature to dissociate nucleoprotein complexes.
Add 0.2 volume of chloroform per 1 volume of TRIzol reagent. Cap the tubes securely and vortex samples vigorously for 15 seconds.
Incubate samples at room temperature for 5 minutes.
Centrifuge the samples at no more than 12,000 x g for 15 minutes at 4°C. The mixture will separate into three phases (Figure 1).
- Lower organic phase.
- Interphase.
- Upper aqueous phases containing RNA.
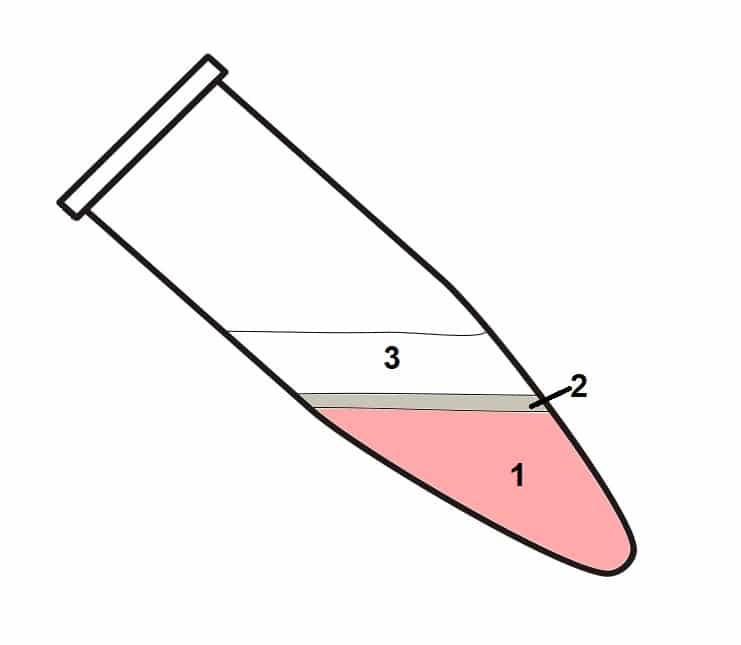
3. RNA Precipitation
Carefully transfer the upper aqueous phase without disturbing the interphase into a fresh tube. The volume of the aqueous phase is usually about 60% of the TRIzol volume used in step 1.
Use 0.5 ml of isopropanol per 1 ml of TRIZOL to precipitate the RNA from the aqueous phase.
Incubate samples at room temperature for 10 minutes and centrifuge at not more than 12,000 x g for 10 minutes at 4°C. The RNA precipitate will form a pellet on the side or bottom of the tube, which can be hard to see by eye.
4. RNA Wash and Resuspension
Remove the supernatant and wash the RNA pellet once with 75% ethanol.
Mix the samples by vortexing and centrifuge at no more than 8000 x g for 5 minutes at 4°C. Repeat the above washing procedure and remove all leftover ethanol.
Air-dry or vacuum dry RNA pellet for 5-10 minutes but don’t heat or centrifuge under vacuum. Also, avoid overdrying the RNA or it will be hard to redissolve the pellet.
Dissolve RNA in DEPC-treated water, by passing the solution a few times through a pipette tip.
5. Determining RNA Concentration and Purity
Once you have your sample redissolved, determine sample concentration and purity by taking OD measurements at 260 nm and 280 nm. The A260/A280 ratio should be above 1.6.
Benefits of TRIzol for Total RNA Extraction
TRIzol has many benefits for total RNA extraction, including:
- Denaturing of RNases.
- Extraction of total RNA, including small molecular weight RNA such as miRNA.
- High-quality RNA.
- Relatively simple to use.
- Allows simultaneous extraction of DNA, protein, and RNA from a sample.
Disadvantages of using TRIzol
TRIzol use in total RNA extraction has some limitations.
- The reagent can be costly compared to other traditional methods of RNA extraction.
- Uses dangerous and hazadous chemicals.
- Extracted RNA can be contaminated with phenol and other contaminants when removing the aqueous layer.
- Can be time-consuming to perform and has a steep learning curve.
Alternative methods to TRIzol for RNA Extraction
The dangerous nature and steep learning curve of TRIzol total RNA extraction have resulted in a market for safer, simpler alternatives for RNA extraction that still provide high-quality RNA.
RNA Extraction Kits
Many commercial kits are available that don’t use TRIzol or other hazardous chemicals. These use spin columns or magnetic beads to capture RNA which can then be eluted. However, they might not be suited to all applications, so check the user manual before purchasing to check that the kit:
- Works with your sample type.
- Isolates RNA you are interested in.
If you are extracting DNA from specialist tissue samples (e.g., blood, fibrous tissue, plant tissue), check to see if there is a kit available. Manufacturers have created and optimized kits for extracting RNA from a range of tissues and source materials.
If you are looking to analyze miRNA or other small molecular weight RNA species, be aware. Many RNA isolation kits, especially those using spin columns, may not successfully isolate these small molecules. However, custom kits designed especially for miRNA and other small RNA molecules are available.
Companies that provide RNA extraction kits include:
- QIAGEN
- ThermoFisher Scientific
- SigmaAldrich
- Zymo Research
- Promega
- Agilent Technologies
- Roche
While RNA extraction kits offer a handy alternative, they can be expensive and if you isolate RNA from various tissues you may need multiple kits.
There are kits that can be used in combination with TRIzol extraction by trapping the RNA following the precipitation step to make subsequent washes and elutions easier and faster. This may be an option if you already have samples stored in TRIzol.
Old-school methods for Total RNA Extraction
If you don’t want to use commercial kits but still want to avoid the cost of TRIzol (and alternatives), consider going back to the old-school methods of total RNA extraction.
Below we share a TRIzol-free method of total RNA extraction from yeast (which can be used for other sources as well) from Vicki Doronina. Note that this method still uses hazardous chemicals, and therefore could be carefully considered before using.
This total RNA extraction protocol uses a mix of phenol and some salts. All that is required is some Tris, SDS, and phenol–chloroform mix. Vicki has never used this protocol on non-yeast cells but she is almost sure that it can be applied to any cell type after the homogenization step in the RNA buffer. Changing the buffer pH from neutral to acidic—pH 4.5—will allow you to isolate aminoacylated tRNAs as well.
Phenol–chloroform RNA Extraction Protocol
1. Grow 25–100 ml of cells to OD600 = 0.25–0.5 (You don’t even need a spectrophotometer for this).
2. Spin cells, wash them in 1 mL dH2O, and transfer to a screw-cap tube. You can snap-freeze pellet at this stage.
3. To 1 volume of cold RNA buffer add SDS to final concentration 0.5% (1/40 volume 20% SDS).
4. Resuspend frozen pellet in 200 µl cold SDS/RNA buffer.
5. Add 1 volume phenol–chloroform; fill with beads to reach the above solution.
6. Break open (usually 30 sec/ 1 min on ice/30 sec on maximum Ribolyser setting) but your routine breakage procedure can be just as effective.
7. Fill tube with RNA buffer (no SDS added), vortex, and spin 5 min keeping cold.
8. You will see phenol–chloroform fraction at the bottom of the tube, white layer of debris, and top buffer level. Take the top level and transfer to a fresh RNase-free Eppendorf.
9. Extract aqueous fraction with phenol–chloroform twice, shaking for 5 min. Add 0.9–1 volume of isopropanol, mix, and spin at RT 15–20 min. Because the buffer contains a lot of salt, no additional sodium acetate is necessary
(If 1/10 3M sodium acetate is accidentally added, get rid of the salts by dissolving the dried pellet in 600 µl RNA buffer, incubating 5 min with shaking, adding 600 µl of isopropanol, and spinning 10–5 min. After this go to step 9).
10. Wash pellet with 70%, air dry, resuspend in 30-50 µl dH2O or TE, and measure OD260/280 ratio.
RNA buffer (50 ml)
- 100 mM EDTA pH 8.0 (10 mL 0.5M stock)
- 100 mM NaCl (1 mL 5M stock)
- 50 mM Tris-HCl pH 8.0 (2.5 mL 1M stock)
- 36.5 mL dH2O
Total RNA Extraction Summarized
There are many methods available for total RNA extraction from a range of samples, from the ‘gold-standard’ TRIzol to commercially available spin kits and old-school phenol–chloroform extraction. Each method of RNA isolation has its pros and cons from the cost, use of hazardous chemicals, and suitability, each of which should be carefully considered before selecting.
Which total RNA extraction method do you use? Leave a comment below.
Originally published March 13, 2013. Reviewed and updated February 2022.







