There are many different types of polymerase chain reaction (PCR) you might have heard of or want to use for a specific requirement in the lab. One of these is touchdown PCR.
To be honest, when I first heard of touchdown PCR, I thought of a landing aircraft, which, as it turns out is not a bad way to think about it.
Touchdown PCR is a very useful technique for improving PCR amplification specificity and sensitivity.
However, using this technique in the lab can be trickier than it might seem at first. So in this article, I’ll provide a primer on touchdown PCR and give you 5 key tips and references for perfecting it.
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
The Problem of Non-Specific Amplification
So what’s the issue with non-specific amplification? PCRs can, and often do, suffer from issues such as non-specific amplification. Mispriming, where primers bind non-specific sequences in the DNA template, resulting in the DNA polymerase amplifying the wrong template DNA sequence, is one cause of non-specific bands on gel following PCR.
How to Eliminate Non-Specific Amplification in PCR
Careful design of PCR primers can help reduce the change of off-target binding, although the sequence of the desired target DNA does limit the choice. Optimizing the PCR conditions by testing various annealing temperatures and other conditions is another way, although this can be time-consuming to complete.
Another, less time- and resource-intensive option, is to use touchdown PCR.
What is Touchdown PCR?
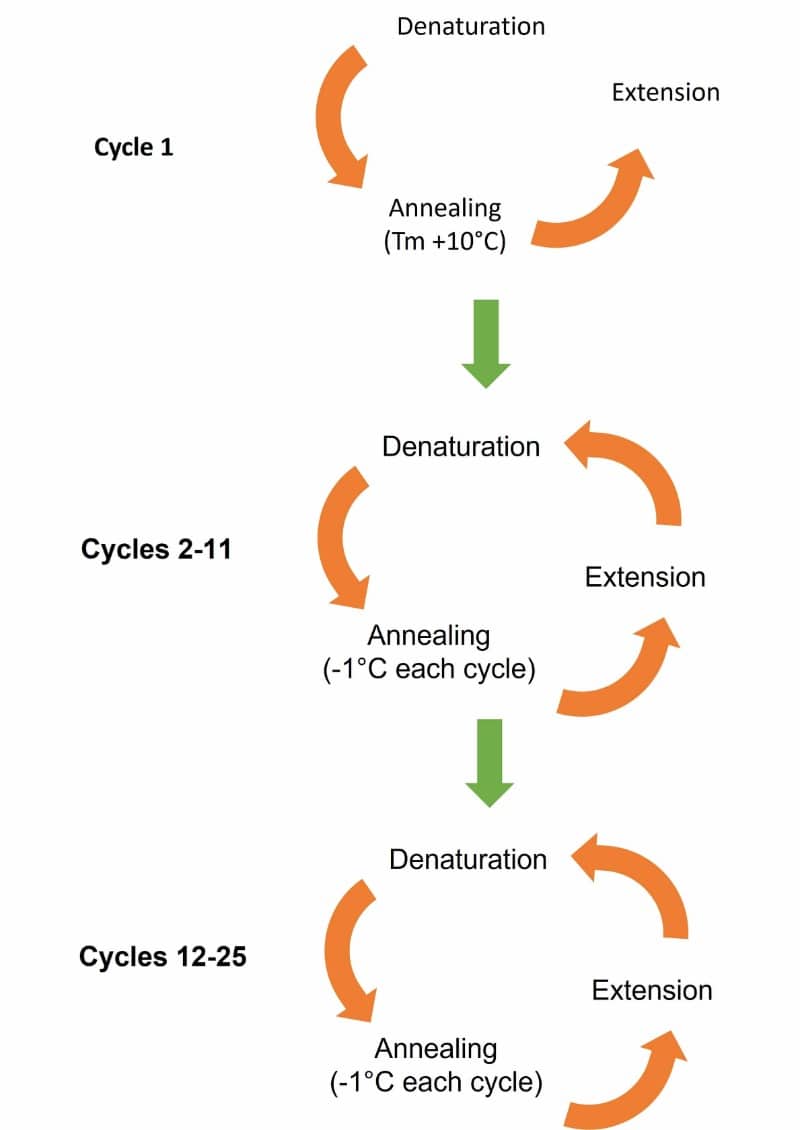
Touchdown PCR is a modification of PCR used to help reduce non-specific annealing and amplification. The initial annealing temperature is set higher than the optimal melting temperature (Tm) of the primers and is then gradually reduced over subsequent cycles until the Tm or “touchdown temperature” is reached, much like the touchdown of an airplane onto the runway (Figure 1).
A gradual lowering of temperature to a more permissive annealing temperature during the course of cycling favors amplification of the desired amplicon.
Advantages of Touchdown PCR
In all types of PCR, it is essential to determine the optimal annealing temperature. This is normally determined based on the Tm of the primer-template pair.
But, primer Tm can be affected by the individual buffer components, and even primer and template concentrations, meaning any calculated primer Tm value is only an approximation. [1]. Therefore, it is often difficult to find the right annealing temperature for a given primer/template combination.
Too low annealing temperatures can lead to primer-dimer formation and non-specific products while too high temperatures can reduce yield due to poor primer annealing.
By using temperatures higher than the calculated Tm in the initial cycles, touchdown PCR favors the accumulation of amplicons whose primer-template complementarity is the highest. The stepwise transition to a lower temperature during the subsequent cycles guards against lower yields by making use of the desired amplicons in the reaction that now outcompete any non-specific products or primer-dimers if present.
Touchdown PCR Protocol
There is a protocol published in Nature Protocols, [2] which works very well and is a great reference to start off with for touchdown PCR.
The suggested cycling program has two phases. The first phase of touchdown programming uses a Tm that is approximately 10°C above the calculated Tm. The temperature is reduced by 1°C every successive cycle until the calculated Tm range is reached. This is done for a total of 10–15 cycles.
Phase 2 follows generic PCR amplification of up to 20–25 cycles using the final annealing temperature reached in the touchdown phase (Table 1).
Table 1: An example of a touchdown PCR protocol based on a Tm of 57°C.
Step | Temperature (°C) | Time (minutes) | Stage and number of cycles |
1. Initial denature | 95 | 3:00 | |
2. Denature | 95 | 0:30 | Stage 1 10 cycles |
3. Anneal | 67 (Tm +10) | 0:45 | |
4. Extensions | 72 | 0:45 | |
5. Denature | 95 | 0:30 | Stage 2 15-20 cycles |
6. Anneal | Last anneal temp -1 | 0:45 | |
7. Extension | 72 | 0:45 | |
8. Final Extension | 72 | 15:00 |
The cycles and temperature drop during the touchdown phase can be adjusted from 1–3 cycles per 1-3°C drop in temperature if non-specific products are still observed or if the yield is low. You can also set the final annealing temperature 1–2°C below the calculated Tm.
Five Key Tips for Successful Touchdown-PCR
1. Keep Reactions Cool
Keeping all reactions cold until thermal cycling starts is crucial to avoiding non-specific priming even with touchdown PCR. So get that ice bucket ready and keep all your components cold until the last second!
2. Use a Hot-Start Setup
Ok! So I know we just said to keep cool. But once your reactions are ready to go, a hot-start setup is preferred to help further reduce non-specific interactions during the initial cycles.
3. Consider Additives
If you are still getting issues when using touchdown PCR, consider combining it with additives. There are several useful PCR additives, which can help boost specificity in your PCR reaction, especially if you have difficult targets (e.g., GC-rich sequences).
4. Keep Cycle Numbers Low
Too many cycles can lead to the appearance of non-specific bands in the gel, indicating non-specific amplification. To limit this, keep the total number of PCR amplification cycles, including the touchdown phase to below 35.
5. Add an Extra Denaturation Cycle
An extra 1 min denaturation cycle at 96°C or 97°C may be extremely useful for difficult templates.
Touchdown PCR Summarized
In short, touchdown PCR is a technique to limit non-specific amplification by starting with a high annealing temperature and slowly decreasing this over PCR cycles until the desired annealing temperature is reached.
You can use touchdown PCR to overcome issues of non-specific amplification and primer dimer formation. So now you understand what touchdown PCR is, what its specific advantages are and you are armed with some key tips to make your reactions go smoothly. We hope this helps- if you’ve had any successes (or disasters!) using touchdown PCR let us know in the comments and share your top tips!
Need help making your PCR fail (a bit) less often? Download our free notorious PCR inhibitors poster and pin it up near your DNA engine. Or download the Bitesize Bio PCR eBook for more comprehensive practical guidance.
References
- Roux K (1995) Optimization and troubleshooting in PCR. PCR Methods Appl. 5 185-94.
- Korbie D Mattick J (2008) Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc 3, 1452–1456.
Originally published July 20, 2009. Updated and republished, July 2022.
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.