Discover what RNA quality control is, why it’s so important for your experiments, and how to undertake it.
After extensive extraction protocols have been carefully followed, you finally have your RNA in hand. Now what? The success of downstream applications, such as microarrays, RNA-seq, and quantitative real-time PCR, relies upon having high-quality, intact RNA.
So it is worth your while to perform some RNA quality control to ensure you have an intact RNA preparation, at the correct concentration and purity, before jumping into your experiments.
What is RNA Quality Control in The Cell?
Firstly, RNA quality control in a mammalian cell can be quite different from what we are referring to here. [1] RNA quality control in a cell is the controlled process by which RNA molecules are actively monitored for integrity. Specific RNA quality control machinery exists to control the destruction of any aberrant RNA molecules. This involves processes such as mRNA 3′-end processing and nonsense-mediated decay. In turn, this protects the cells against misfolded proteins being produced.
However, we are focusing here on how to perform good RNA quality control in the lab, not the cell! There are three key things you want to check your RNA in the lab for: quantity, purity, and integrity. We’ll walk you through each of them below.
1. Measuring RNA Quantity
The traditional and least expensive way to determine RNA quantity is to measure UV absorption of the sample using a spectrophotometer.
A260 for quantification: RNA has a maximum absorption at a wavelength of 260nm. Therefore your RNA concentration is determined by the absorbance reading at 260nm as given by the following conversion using the Beer Lambert Law: an A260 of 1.0 is equivalent to 40 µg/mL of RNA.
2. Measuring the Purity of Your RNA Samples
To measure the purity of the RNA in your samples, you need to assess what else might have carried over from your RNA prep. It’s important to check the absorbance of your samples at different wavelengths corresponding to other biological molecules, such as proteins.
Measuring A280 for Protein Contamination
In addition to the A260, measurements should also be taken at 280nm. The A260/A280 ratio is an indication of the level of protein contamination in the sample. Pure RNA has an A260/A280 ratio of 2.1. However, values between 1.8–2.0 are considered acceptable for many protocols.
A230 for Other Contaminants
RNA preparations can also contain contaminants such as guanidine salts and phenol (commonly used in RNA isolation protocols). A high peak at A230 indicates contamination with either of these. The ideal A260/A230 ratio is greater than 1.5.
A Note on the NanoDrop™
One significant disadvantage of using traditional spectrophotometers for these measurements is that they will require a large sample volume. Typically, spectrophotometers will require a sample volume of at least 200 µL, depending upon the cuvette used.
However, the NanoDrop™ is an excellent spectrophotometer for measuring RNA levels, requiring only 1–2 µL of a sample. Furthermore, the NanoDrop also contains preset programs for RNA measurements that automatically read the OD at 230, 260, and 280 and calculate the concentration of the RNA and A260/A280 and A260/A230 ratios.
Many departments and individual labs now contain the NanoDrop or similar instruments, so chat with your lab mates across the hall to see if you can use theirs perhaps!
Three Words of Caution When Measuring RNA Purity
- UV absorbance measurements can change depending on the pH of the RNA solution. The best results are obtained when RNA is solubilized in TE buffer.
- Always blank the spectrophotometer with the same solution in which the RNA is diluted.
- And remember that RNA concentrations below 20 µg/mL may not give reliable readings.
3. Measuring the Integrity of Your RNA Samples
So if you’ve measured the quantity and purity of your RNA samples and got decent numbers, you’re good to go, right? Unfortunately, this might not be the case. Your RNA may have been degraded!
RNA degradation can happen in the lab for a whole host of reasons. Degraded RNA doesn’t perform well in downstream applications. Therefore it is important to check the integrity of your RNA preparation.
The Cheap and Cheerful Approach To Measuring RNA Integrity: An Agarose Gel
The least expensive method for checking RNA integrity is to run the RNA on a 1% standard agarose gel and examine the ribosomal RNA (rRNA) bands (formaldehyde gels are not required for this quick assessment). To do this, you need to:
- Make a 1% agarose gel plus nucleic acid stain of your choice and allow it to set.
- Mix a small volume of your RNA prep with a loading dye.
- Add the RNA prep and loading dye into a well.
- Add your running buffer and run the gel as usual.
- Image the gel.
Figure 1 shows a range of results you might get from running your RNA samples on an agarose gel. The upper ribosomal band (28S in eukaryotic cells and 23S in bacterial cells) should be about twice the intensity of the lower band (18S in eukaryotic cells and 16S in bacterial cells) and should be crisp and tight, as shown in Figure 1 in Lane 1 of the gel.
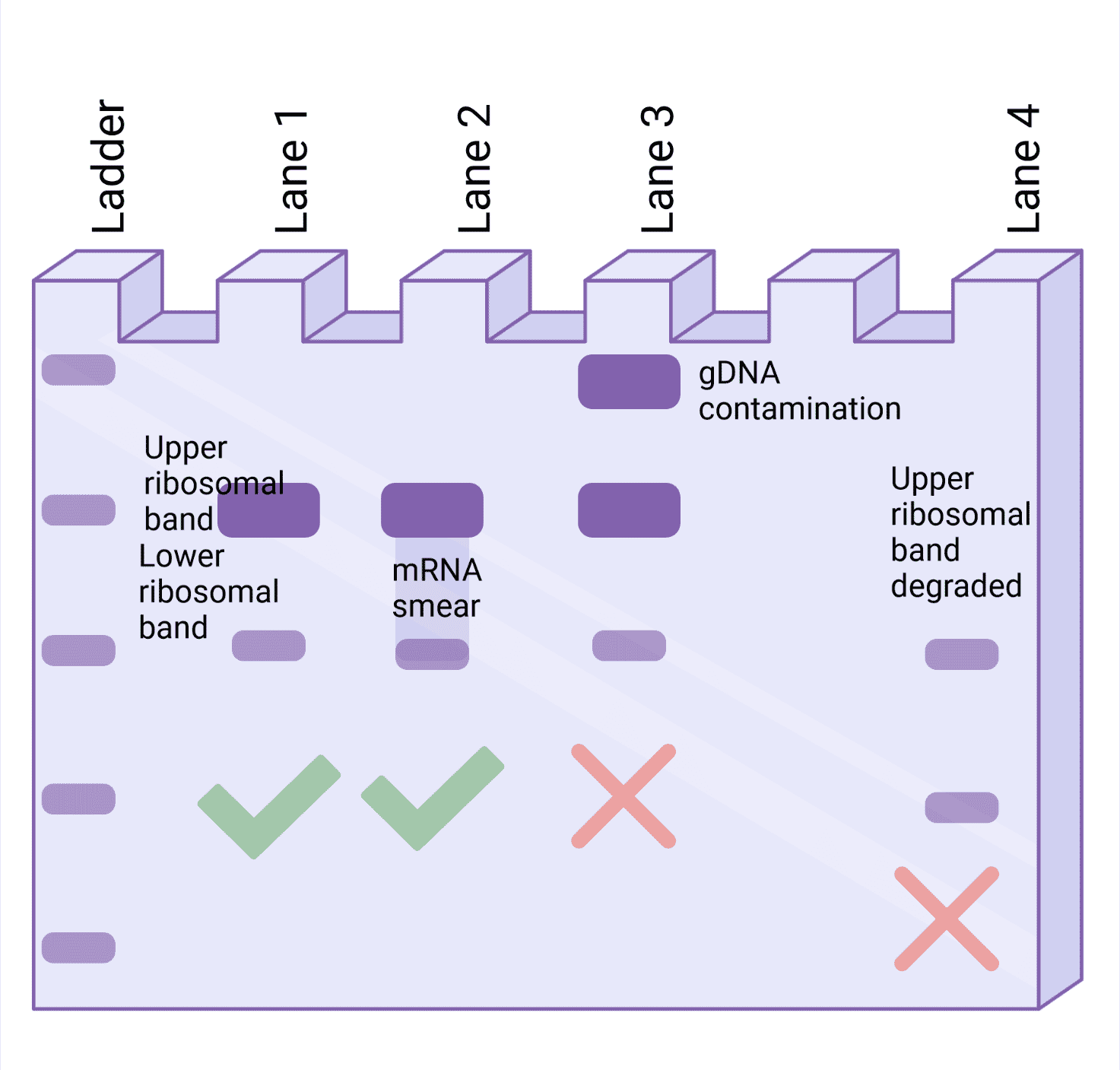
However, if the rRNA bands are of equal intensity, it suggests some degradation has occurred. mRNA runs between the two ribosomal bands and might be seen as a smear, as shown in Lane 2 of the example gel in Figure 1. Don’t panic, this is ok!
If you see a significant large-sized band corresponding to genomic DNA (gDNA) that runs much higher up the gel due to its large size, this strongly suggests that the RNA is contaminated with DNA, as depicted in Lane 3 in Figure 1. Furthermore, smearing below the rRNA bands suggests that you have poor-quality RNA, as shown in Lane 4 of Figure 1.
However, just like using a spectrophotometer to assess the RNA quantity, one potential issue with using agarose gels to assess the integrity of RNA is the amount of RNA required typically for good visualization. Therefore you might want to consider a different option.
A More Expensive Option For Measuring RNA Integrity: Bioanalyzers
A second method to check the integrity of your RNA is to use a bioanalyzer, such as the Agilent Bioanalyzer. Bioanalyzers use small amounts of RNA (1–2µL) and microfluidics to determine the quantity and quality of RNA samples. The analyzer measures the sizes of the rRNA bands and determines an RNA Integrity Number (RIN) to standardize between RNA samples. [2] Bioanalyzers are expensive but can often be found in core facilities and used for a fee.
What to Do If Your RNA Is Degraded?
If you find that your RNA is degraded, it is definitely worth the investment to prepare new RNA before performing experiments, making sure you take measures, paranoid or otherwise, to prevent RNAse contamination!
RNA Quality Control Summarized
It’s important to check the quality of your RNA before you use it in downstream experiments as the concentration might be too low, there could be contamination with protein or reagents from the extraction process such as phenol, or it may have been degraded.
Originally published in March 2014. Reviewed and updated in September 2022.
References
- Meenakshi K and Parker R (2007) RNA Quality Control in Eukaryotes, Cell, 131, 4, 660-668.
- Schroeder, A., Mueller, O., Stocker, S. et al. (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biol 7, 3.