How do you win a Nobel Prize?
A good deal of brainpower, definitely. A dash of foresight and bravery to stake your research career on a legitimate frontier helps. A splash of insight to ask the correct questions at the risk of ridicule is a must.
Serendipity.
Cryo-Electron Microscopy (cryo-EM) entered the mainstream, seemingly from nowhere, when it won the Nobel Prize in Chemistry in 2017.
Furthermore, the choppy blend of questions, results and timing that chart its rise makes for an intriguing tale.
So let’s take a step back. Enjoy a short history of cryo-electron microscopy.
The Power of an Image
A black hole. King Tut’s death mask. Ernest Shackleton’s sunken ship, Endurance.
What do all these have in common?
That’s right—they’ve all been photographed.
The advances in any discipline thanks to an image are impossible to understate. This is self-evident. Hence, in part, the monumental efforts undertook to image the objects listed above amongst many others.
The same reasoning applies to the atomic world.
For over a century, X-ray crystallography has been the dominant technique used to capture images of molecules. The enormous scientific contribution forged by it is reflected by the number of Nobel Prizes awarded to its vanguard. Approximately 15. [1]
The Limitations of X-ray Crystallography
Living organisms are dynamic; a reductive approach to solving their components piece-by-piece will never grant a complete understanding of them.
The limitations of capturing biological processes have been appreciated for decades. They necessitate an imaging technique amenable to large and dynamic samples.
Enter cryo-electron microscopy.
A Short History of Cryo-Electron Microscopy
The slow, inching progress of cryo-EM towards the scientific mainstream can be told as a story with three parts:
- Three brilliant scientists successfully conducted fundamental research.
- A struggle against the benchmark technique, X-ray crystallography.
- Landmark results in cryo-EM herald an attractive, mainstream method.
If you’re in a massive rush and don’t have time for a story, check out the graphical timeline below in Figure 1.
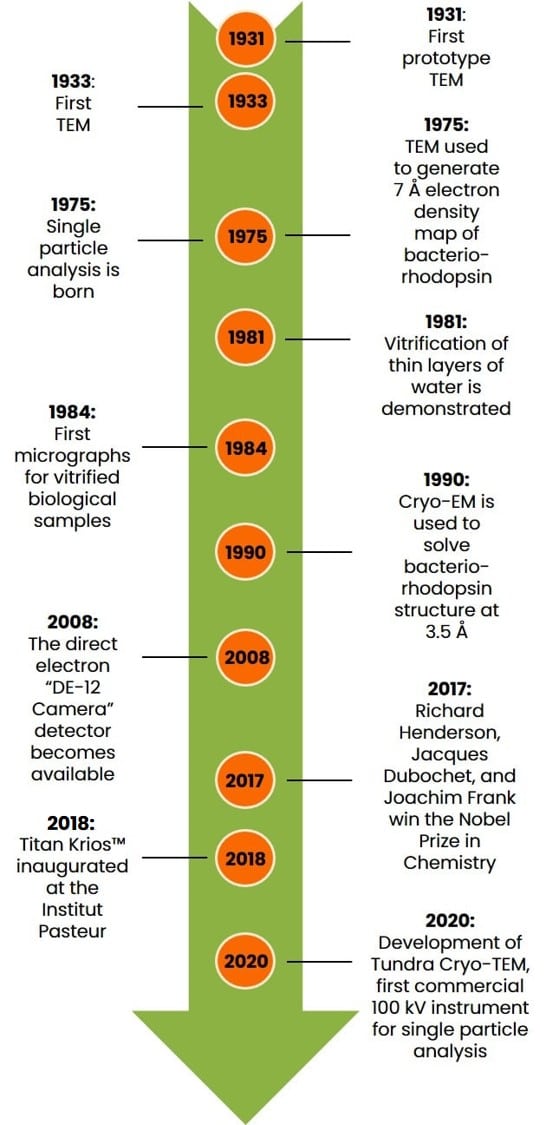
Otherwise, Let’s begin.
1. Early Hurdles: Resolution, Radiation Damage, and Computation
A landmark paper published in Nature in 1975 by Richard Henderson (who would later become one of three recipients to share the 2017 Nobel Prize in Chemistry) and Nigel Unwin showcased a 7 Å electron density map of the membrane protein bacteriorhodopsin obtained via electron microscopy. [2]
In addition to proving that transmembrane helices exist and showing membrane proteins had explicit structures, it demonstrated unequivocally the efficacy of using electron microscopy to study biomolecules.
However, in 1975 X-ray crystallography enabled protein structures to be solved at < 3 Å, although in no way near the same abundance as we see them published today
High-resolution images of molecules reconstructed from electron micrographs necessitated, due to low signal-to-noise, electron detectors of a type that simply didn’t exist back then.
Increasing the dose of electrons wasn’t a feasible solution because it damaged the delicate biological samples beyond use.
So chasing resolution was, in effect, chasing your tail. What was needed was another watershed discovery.
It came from an oblique angle. Solid-state physics.
Freezing Water Without Forming Ice: Vitrification
Vitrification of water is the process of cooling it so rapidly that it becomes a solid without forming ice. Slowly cooling water in the presence of biological specimens harms and denatures them. This would obviously ruin cryo-EM samples. So, you must vitrify them.
The problem is, it’s actually quite challenging to do. Doubly so when the desired goal is an ultrathin layer of vitrified water that encapsulates delicate biomolecules in their native state.
Vitrifying biological samples increases the resolution to which data can be collected through the following three ways:
- Reducing the radiation damage caused by incident electrons.
- Protecting samples from the harsh vacuum inside the microscope.
- Locking water molecules in place around native samples.
Remember that water molecules are part of a sample’s quaternary structure.
Two Critical Discoveries Come in Quick Succession
In 1981, a paper published by Jacques Dubochet and members of his lab demonstrated a technique to produce sub 1 µm thick vitrified water using liquid ethane (recall the importance of sample thickness in electron microscopy). [3]
Then, in 1984, the same group published a paper exhibiting cryo-electron micrographs of single, undamaged virus particles embedded within a layer of vitrified water (also called amorphous ice). [4]
For these efforts, Jacques Dubochet also shared the 2017 Nobel Prize.
That’s two pieces of the puzzle:
- Electron micrographs can be used to construct electron density maps of biomolecules.
- Cryogenic techniques can be harnessed to embed and preserve delicate samples.
But how are high-resolution electron density maps produced from noisy electron/cryo-electron micrographs?
Single Particle Analysis
That accolade goes to Joachim Frank, who in 1975 solely published the unassumingly titled paper: “Averaging of low exposure electron micrographs of non-periodic objects.” [5] Or single particle analysis as it’s now known.
This study provided the framework for taking noisy microscopy data for massive numbers of non-crystalline biomolecules and averaging them to maximize the usable data and ultimately translate it into electron density maps.
Maps that are just like those obtained using X-ray diffraction.
For this work, he further shared the 2017 Nobel Prize in Chemistry.
And there you have it. Three separate feats of Nobel-worthy science ready to revolutionize molecular biology just like PCR was to do shortly afterward. [6]
And then, seemingly, nothing happened. In fact, cryo-EM was derided as “blobology” due to the comparatively low-resolution data it provided. [7]
2. Into the Mainstream: Cryo-Electron Microscopy Wins the Nobel Prize
Okay, so it’s not as if the technique was abandoned it just didn’t take off either. In fact, it was to be a long time before it was to do so. There are several reasons why:
Timing Is Everything
The first is an issue of timing. We have already mentioned X-ray crystallography, and I alluded to the PCR revolution, which widened the bottleneck of sample crystallization.
Won’t your sample crystallize? Okay, tether it to a crystallizable chaperone, chop it down into a single domain, mutate flexible loops, turn it into a chimera, make a library of all these and try to crystallize them all.
Plasmids and recombinant DNA technology also enabled samples for X-ray crystallography to be produced using the expression systems we take for granted today. Previously, samples were extracted from large quantities of natural sources. [8] This is a messy, probably quite smelly, inefficient, and limited way of doing things.
Inadequate Electron Detectors
The second is an issue of hardware. Specifically, electron detectors. You see, poor signal-to-noise ratios and low contrast micrographs are substantial issues in cryo-EM. To generate electron density maps adequate for model building at resolutions the research community cares about, you really need to wring out as much usable data as possible.
So pivotal is this problem that Richard Henderson attributes the enormous increase in high-resolution cryo-EM structures to the introduction of direct electron detectors in 2008. [9] These detectors can deal with systematic noise and produce clearer micrographs, making particles easier to classify.
Insufficient Computing Power
The third issue is computing power. Or lack of computing power, to be precise. Generating electron density maps from thousands upon thousands of grainy images of particles takes a lot of computing power.
Exactly how much computing power depends on several factors, including the number of particles being analyzed, their dimensions, symmetry, and the resolution to which the data is collected. [10]
Even today, in an era in which our smartphones contain multicore processors faster than PCs from ten years ago, solving structures via cryo-EM takes a lot of computing power.
Electron density maps constructed using single-particle analysis may take thousands of hours of processor time. Plus, it can be expensive to tap into supercomputer clusters.
Small wonder it took decades to catch on.
Catch on it did, however, and it’s not hard to see why. Visit the Electron Microscopy Data Bank (EMDB) and choose a year. I chose 2015 because it’s recent but before the Nobel Prize was awarded.
Of the 640 depositions that year, 90 correspond to viruses, 40 correspond to cellular components, including parts of a flagellar motor, and 105 correspond to samples weighing > 1 MDa.
The top three journals that year were Science (75 publications), Nature (66 publications), and PNAS (53 publications).
3. The Revolution: Modern Applications of Cryo-Electron Microscopy
So, why did cryo-electron microscopy win the Nobel Prize?
Well, take a look! Here are eleven incredible structures that cryo-EM has enabled (Table 1).
Structure | Year | Size | Sample Type |
1990 | 27 kDa | Membrane protein | |
2020 | 88 kDa | Membrane protein | |
1999 | 120 kDa | Virus | |
2020 | 190 kDa | Virus | |
2022 | 2.7 MDa | Tissue | |
2022 | 1.1 MDa | Tissue | |
2020 | 5.0 MDa | Organelle | |
2021 | 2.0 MDa | Organelle | |
2021 | Unavailable | Organelle | |
2021 | 1 MDa | Filament | |
2022 | 10 kDa | Nuclear receptor |
Note that for the membrane protein embedded in a liposome, the corresponding size doesn’t include the liposome. For virus samples comprised of identical polypeptide chains, sizes correspond to their atomically unique portion. Tissue sizes are based on the estimated molecular weights of their constitutive proteins. The size of the flagellar MS ring is inferred from 34 copies of 60 kDa FliF subunits. The bovine tracheal cilia were isolated from a natural source, so a molecular weight is unavailable.
And for your viewing pleasure, I’ve rendered images of some of those structures. Check them out in Figure 2 below.
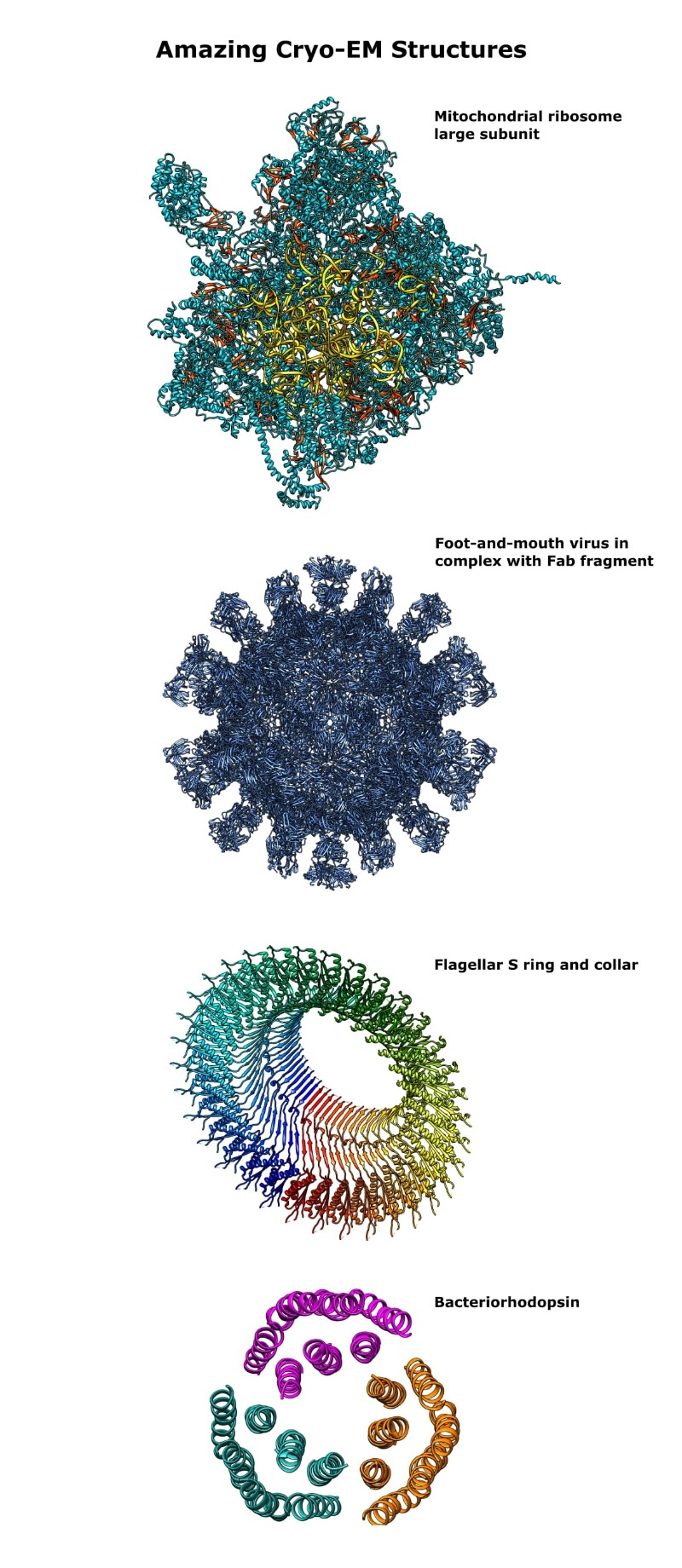
As of 2021, there is a cryo-EM structure of an entire flagellar motor, [11] but my rough-and-ready image-making skills would not do it justice!
If that didn’t convince you, check out the structures related to COVID-19 and see how many were solved using electron microscopy. (I’m sorry, but it’s only a matter of time before that specter showed up.)
Plus, the vertiginous rise in interest in the technique means that you can now partner with external agencies who support you with a cryo-EM project!
The End
For now, at least.
If ever there was an example of the benefits of cross-disciplinary thinking, you’ve just read it!
We’ve touched on solid-state physics, computer science, and image and signal processing, all in the pursuit of a molecular biological goal.
Brains, timing, and good old-fashioned luck have played a part in the rise of cryo-EM.
While realistically, few of us can hope to win a Nobel Prize, we could all perhaps benefit from looking above the parapets of our own particular disciplines.
If you enjoyed this short history of cryo-electron microscopy, get in touch! I’m sure you all have loads of insights to add.
Looking to bring cryo-EM to your research? Get started by exploring the different techniques and applications in cryo-EM.
References
- Galli S (2014) X-ray Crystallography: One Century of Nobel Prizes. J Chem Educ 91:2009–12
- Henderson R and Unwin PN (1975) Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257:28–32
- Dubochet J and McDowall A (1981) Vitrification of pure water for electron microscopy. J Microsc 124:3–4
- Adrian M, Dubochet J, and Lepault J et al. (1984) Cryo-electron microscopy of viruses. Nature 308:32–6
- Frank J (1975) Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy 1:159–62
- Zhu Het et al. (2020) PCR past, present and future. Biotechniques 69:317–25
- Saibil HR (2017) Blob-ology and biology of cryo-EM: an interview with Helen Saibil. BMC Biol 15:77
- Dauter Z and Wlodawer A (2016) Progress in protein crystallography. Protein Pept Lett 23:20110
- Monya B (2018) Cryo-electron microscopy shapes up. Nature 561:565–67
- Sigworth F (2016) Principles of cryo-EM single-particle image processing. Microscopy 65:5767
- Tan J, Zhang X, and Wang X (2021) Structural basis of assembly and torque transmission of the bacterial flagellar motor. Cell 184:2665–79








