Probing Nucleic Acid-Protein Interactions with EMSA
The electrophoretic mobility shift assay (EMSA) is a powerful technique for detecting specific-binding of nucleic acid-protein complexes. Over the past 30 years, EMSA has been the “go to assay” to investigate the qualitative interactions between nucleic acids (DNA or RNA) and nucleic-acid binding proteins. Through the use of radio-labeled probes, one can compare the location of the signal on the polyacrylamide gel to determine the qualitative interaction. In addition, with new advances in computer and optical imaging technologies, EMSA has transitioned its role from merely qualitative to a quantitative assay.
Shifting Our Understanding of EMSA
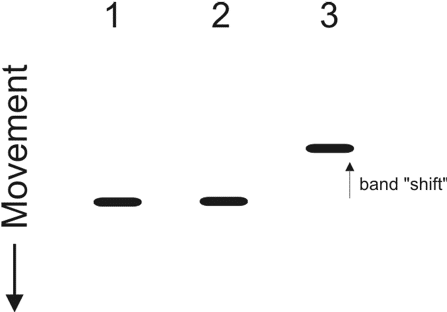
Gel electrophoresis is a commonly used method to separate a group of nucleic acids or proteins based on molecular weight. Typically, an agarose gel is used when running nucleic acids. On the other hand, polyacrylamide gel is often used for the separation of proteins. EMSA takes advantage of the fact that under the condition of an even electric field in gel electrophoresis, large nucleic acid-protein complexes will migrate much slower than a singular protein (Figure 1). To explain this, let’s look at protein migration. Say protein x is a basic unit (monomer) and it is able to associate with other protein x’s to form a larger complex called tetramer. The tetramer is always going to run much slower when compared to a monomer in a gel electrophoresis experiment under the same electric field.
If you apply this same principle to a DNA-binding protein, you would expect a shift in band migration following gel electrophoresis if specific DNA-protein complexes are present. Band migration of DNA-protein complexes is normally detected with radio-labeled DNA probes to detect a sequence-specific interaction. These probes typically consist of a linear stretch of binding sequences that you are examining. You can choose to label either the front (5’) or the back (3’)-end of the linear probe with radio-active isotopes such as phosphate-32 (32P). Following electrophoresis, the gel is visualized by exposure to an x-ray film. To be even more sure that a specific protein is binding to the DNA, researchers can also add monoclonal antibodies specific for the nucleic acid-binding protein in the complex, resulting in a “gel super-shift”.
By the way, if you are still confused about how to run a protein-separation gel, check out the following articles:
- How SDS-PAGE Works(https://bitesizebio.com/580/how-sds-page-works/)
- The Nature of Denaturing (Protein Gels, that is!) (https://bitesizebio.com/20794/the-nature-of-denaturing-protein-gels-that-is/)
There are alternative methods to study protein-DNA interactions. For example, if you want to characterize the DNA-protein complex, one can employ immunoprecipitation (or DNA pulldown), which involves using secondary antibody labeled with magnetic beads. The samples can be purified (pulled down) by passing through a magnetic column. In addition, a more specialized chromatin-immunoprecipitation assay (ChIP) can sometimes be substituted for EMSA.
What Has Changed Over the Years?
A. Type of Detection Methods
- Radioactive Probes Vs. Fluorochrome Probes. As I have mentioned in the previous paragraph, radio-labeled nucleic acid probes were used for EMSA detection in the old days. Typically, this involves either a 5’ (front) or 3’ (back) end labeling with phosphate-32. Although the principle mechanism behind the gel-shift assay stays the same, new reagents for detection have been developed. Nowadays, we need to stay away from radioactive materials that could harm ourselves and contaminate our environment. Instead, researchers use fluorochrome-based dyes or luciferase-based light-emitting systems. Not only are these new systems safer in terms of our health and the environment, but they are also more precise than radioactive setups, and tend to generate less background noise. To use fluorochrome probes, you need an optical image capture system that has an excitation light source (i.e laser). Typically, these imaging apparatus are made of two parts: 1) a digital camera system to control the focus and exposure of an image; and 2) wide spectrums of light sources (from ultra-violet to lasers with different excitation wavelengths) to complement the different variety of fluorochromes in the market. With proper technique, you can obtain great results with this imaging system.
- Chemiluminescent EMSA. Typically, this kind of reagent kit provides biotin-labeled controlled DNA and nuclear extract control sets for you to standardize your experiment. The chemiluminescence reaction provides a clear and strong signal if you use the proper detecting equipment. The slight downside is that you will need to biotinylate your own DNA template (target of interest). But don’t you worry: you can usually get your DNA oligos customized from a commercial source.
- SYBR Green nuclear stain & protein red stain. Several labs have successfully used commercial reagent kits for EMSA (1,2). These kits are designed to separate nucleic acids and proteins by color. The nucleic acids will be stained green and the proteins will be stained as red. Thus, when the two forms a complex, the final colour would be yellow. Again, you can visualize your result on a commercial optical imaging system. This is a pretty neat assay for a quick easy test, especially when labeling of the target nucleic acids is not a best option.
B. Other Methods to Detect Protein Nucleic Acid Complexes
- Rapid agarose gel electrophoresis. A research group led by Lewis KA has developed a new method using agarose gel (3), which is much easier to make and perform than the polyacrylamide used with traditional SDS-PAGE. Moreover, the group is able to reduce the runtime and still obtain high-resolution data by running at high voltage, which is in contrary of the current wisdom on gel electrophoresis 101. Typically, increasing the voltage could lead to a smearing effect.
- Microfluidic Vs. 96-well Plate Microarrays. There are also a couple of more specialized assays. For example, NF-kB (Nuclear Factor kappa-light-chain-enchancer of activated B cells) detection kits use immobilized target-sequence DNA for high throughput screening (4). It works nicely in a 96-well plate format. Your sample nuclear extracts can be added and a positive result will be detected using a monoclonal antibody capable of detecting the nuclear-DNA complex.
Another potentially high throughput method involves the use of microfluidic chips that are designed for the separation of differently sized DNA fragments. A research group led by Smith et. al reports that the DNA-protein complexes can also be separated using these microfluidic devices (5).
Conclusions:
EMSA has such a broad applications on DNA-protein interactions and different protocols are still being added and modified. I can’t wait to hear more about how EMSA continues to develop!
Have you used EMSA? Share your experiences with us by writing in the comments section!
References
- Keyhani et al. Interaction between TATA-Binding protein(TBP) and Multiprotein bridging factor-1 (MBF-1) from the Filamentous insect pathogenic fungus Beauveria bassiana. PLoS ONE. 2015 Oct 14; 10(10): e0140538. https://doi.org/10.1371/journal.pone.0140538
- Patton WF et al. The utility of a two-color fluroscence electrophoretic mobility shift assay procedure for the analysis of DNA replication complexes. 2004 Aug; 25(15: 2439-46. .https://www.ncbi.nlm.nih.gov/pubmed/15300760
- Ream JA, et al. Rapid agarose gel electrophoretic mobility shift assay for quantitating protein: RNA interactions. Anal Biochem. 2016 Oct 15; 511: 36-41.
- Piper J. (2011, March). Paradigm Gel Shift [web log post]. Retrieved March 2019 from https://www.the-scientist.com/technology/paradigm-gel-shift-54966
- Clark J, et al. Mobility-shift analysis with microfluidics chips. Biotechniques. 2003 Sep; 35(3): 548-54.







