Have you ever wished you could snag individual strands of DNA or RNA with a lasso? Or look at them one by one, figuring out exactly where they are or what they are doing? Fortunately, there are techniques that exist to label nucleic acids for their visualization and purification! Nucleic acids can be labeled at their 5´ end, 3´ end, or throughout the molecule depending on the particular application, including:
- to generate information on gene integrity and copy number (blot)
- to diagnose specific sequences and chromosomal aberrations (in situ hybridization)
- to simultaneously measure the relative expression of RNAs (microarray analysis)
- to discover protein-nucleic acid interactions (electrophoretic mobility shift assays or FRET)
This article highlights the two main classes of nucleic acid labels available (radioisotope and non-radioactive), and discusses the pros and cons of each.
Labels, Labels, Everywhere
Generally speaking, there are two types of nucleic acid labeling techniques: radioisotope labeling and non-radioactive labeling.
- Radioisotope labeling: Considered as a conventional method for nucleic acid labeling, radiolabeled nucleotides are synthesized using ATP-gamma-32P or 35P. They are easily incorporated into nucleic acid sequences by traditional enzymatic means or by an organism of interest.
Radioactive nucleotides were first used in 1935 by George de Hevesy (Nobel Laureate in Chemistry, 1943) to reveal components of metabolism in rats. Since then, radioisotope labeling of nucleotides has been used in many studies and clinical applications, such as investigations into bacteriophage replication and clinical diagnosis of cancers.
There is a long history of radiolabeling for DNA and RNA applications. While this technique is relatively less expensive than non-radioactive labeling and is widely used, it is important to review the safety concerns of working with radioactive nucleotides in the lab. Furthermore, if you’re looking to get single-molecule level precision in your application, you may want to consider non-radioactive labeling.
- Non-radioactive (chemical) labeling: Nowadays, non-radioactive nucleotide labels are more extensively used due to their relative speed, sensitivity, safety, and versatility. The most common labels are fluorescent ‘tags’ that are synthesized and incorporated into oligonucleotides, but you can also attach a variety of other molecules or proteins to chemically reactive groups like biotin, streptavidin, or fluorophores. Pre-labeled oligos are available from most oligo suppliers like as IDT DNA or Genewiz.
Non-radioactive labels for DNA and RNA are widely used in molecular biology labs. Fluorescent and reactive labels help researchers investigate proteins that interact with nucleotides at a single molecule level (e.g. FRET). Check out the table below summarizing chemical methods for nucleic acid labeling!
While slightly more expensive than radiolabeling, fluorescent and chemically labeled nucleotides and oligos are easy to use for a variety of applications in the lab. Plus, this class of modified nucleotides don’t require extensive training in radioactive isotope handling.
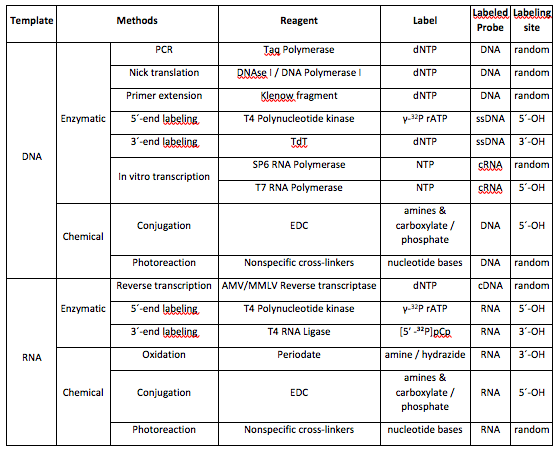
Sources: Jena Bioscience GmbH, Thermo Fisher Scientific, and New England Biolabs
* dNTP or NTP may be labelled with radioactive phosphate, hapten, or fluorophore.
Choosing a Labeling Method
When choosing a labeling system, consider the size and type of nucleic acid you’re working with. Large DNA, plasmid DNA, and RNA for blots and in situ hybridization can be labeled throughout by random incorporation of a covalently coupled label. While covalent probes produce excellent sensitivity, enzymatic methods are more economically convenient in the lab and can label copies of the sample (in PCR labeling). For shorter sequences like oligos, it may be more convenient to order a pre-labeled sample through a company that specializes in labeled DNA, like IDT or Genewiz.
You should also think about the application you’ll be using your labeled nucleic acid in. For protein interaction studies, for instance, large fluorophore labels can interrupt interactions due to their steric hindrance and chemical properties. You can avoid this concern by using smaller labels or by labeling a site that is farther away from the interaction location. If you’re looking for signal amplification, secondary reporters such as antibody conjugates like avidin/streptavidin are an excellent choice.
What nucleic labeling techniques and reporters have you used? Tell us in the comments below!
References
- Ligasová, Koberna. DNA Replication: From Radioisotopes to Click Chemistry. Molecules. 2018 Nov 17;23(11). doi: 10.3390/molecules23113007.
- Jena Bioscience. https://www.jenabioscience.com/
- Thermo Fisher Scientific Nucleic Acid Labeling Brochure. https://www.thermofisher.com/us/en/home/life-science/dna-rna-purification-analysis/nucleic-acid-labeling.html
- New England Biolabs DNA and RNA Labeling. https://www.neb.com/products/dna-modifying-enzymes-and-cloning-technologies/dna-labeling/dna-labeling