Allele-specific expression can occur for various biological reasons, such as gene imprinting, or differential transcription caused by mutations, or single nucleotide polymorphisms (SNPs), or epigenetic alterations. Traditional end-point RT-PCR or qRT-PCR-based methods only detect overall levels of mRNA expression from a given gene rather than mRNA transcripts originating from individuals.
If your project requires more in-depth analysis of allele-specific expression, then some simple modifications to your PCR experiments and protocols can help you to achieve this, either quantitatively or semi-quantitatively. Read on to find out how!
Allele-Specific (AS) Primer Design and PCR
Designing primers for allele-specific mRNA expression is not as difficult as it may sound. The procedure is essentially the same as designing primers to amplify any genomic or transcriptomic region, except that the primer must physically cover the nucleotide(s) of the SNP, indel or mutated base.
For example, if you want to investigate a C-T SNP, then you simply design two different forward primers: one containing the C allele at the 3′ end of the primer, and the other containing the T allele at the 3′ end. Then, design one common reverse primer, so you have three primers in total (see Figure 1).
For this method, although you have to perform two PCRs, one for each allele, you get double the detail! If you want to make this semi-quantitative, you can optimize the number of PCR cycles to capture your products in the linear phase of amplification. Then run your samples on an agarose gel and voila! You can find more information of PCR and primer design in this eBook. For allele-specific design check out WASP and BatchPrimer3.
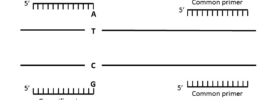
Allele-Specific (AS) Primer Design and PCR – Simplified!
Armed with your allele-specific primers as described above, you can amplify your genes and alleles of choice and run your products on an agarose gel to confirm expression of one, both or neither allele. One interesting and helpful adaptation to this protocol is described in a methods paper by Gaudet et al., (2009)1. They describe how two slight adaptations of allele-specific primers can allow you to amplify both alleles separately but in one PCR. Bingo!
To do this, simply create a mismatch in the nucleotide immediately 5′ of the SNP in the allele-specific primers (see Figure 2). This reduces the chances of the wrong primer binding to the wrong allele and makes your primers a lot more selective. This means that you can use both primers in the same PCR.
Another slight modification that you need to make is to elongate one of the primers. This allows you to distinguish between alleles when you run the products on a gel. Elongating one primer by 10 bases or more is enough to distinguish your products on a 3% agarose gel. The band corresponding to the allele amplified with the longer primer will have a larger product than the band for the allele amplified with the short primer. Again, you can optimize the number of PCR cycles to make this set-up semi-quantitative.

Allele-Specific Gene Expression Using qPCR
With access to qPCR reagents and equipment, you can take a more quantitative approach to allele-specific expression analysis. The qPCR approach is similar to the methods described above in that you amplify the region around your SNP or mutation, but allele-specific probes are used to discriminate the PCR products, with different fluorophores for each allele.
Generally, this is a more costly approach but with the benefit that someone else has done the optimization steps for you! qPCR-based approaches are ideal for quantifying allelic expression when you want to measure the output of both alleles, for example in eQTL analysis, rather than going for the ‘yes or no’ answer that you can obtain with the semi-quantitative protocols described above.
These qPCR-based methods are also based on the SNP genotyping assays described elsewhere on BitesizeBio2,3. An alternative qPCR-based approach would be to amplify your region of interest with SYBR green before performing a melt curve analysis to differentiate your products. An excellent example of this can be found in this article by Dhas et al. This is another viable option however both approaches should be validated by creating a standard curve (see below).
Using Pyrosequencing
Pyrosequencing is an effective tool for detecting and quantifying allele-specific expression and for directly measuring the ratio of expression of one allele relative to another. This, again, is very useful for expression quantitative trail locus eQTL analyses in order to quantify allelic expression imbalance.
You can check out our article covering a basic introduction to pyrosequencing. Firstly, you would amplify your region of interest with your SNP or mutation in the middle of the amplicon. This PCR step needs to performed using two primers, one of which contains a 5′ biotin tag. Then, bind your PCR product with streptavidin beads, wash, and denature. Next, incubate with a sequencing primer that binds upstream of your SNP or mutation. A DNA polymerase reaction elongates along the strand to cover your nucleotide of interest. The data from your sequencing reaction will then allow you to quantify how much of each allele is present in your sample.
Standard Curve
For the more quantitative qPCR and pyrosequencing approaches, it is good scientific practice to test your assays before applying them to your samples. You need to ensure that:
1. The assay is capable of detecting both alleles
2. The assay can detect both alleles equally, or if not, then you need to know whether or not there is a bias
3. The assay is capable of detecting changes in expression, and you know the limits of detection
Create a standard curve by amplifying DNA or cDNA from, for example, major and minor allele homozygous samples. Then, create mixes of DNA to test your assay across a range of allelic percentages, for example:
1. 100 % major allele, 0 % minor allele
2. 80% major allele, 20 % minor allele
3. 60 % major allele, 40 % minor allele
4. 40 % major allele, 60 % minor allele
5. 20 % major allele, 80 % minor allele
6. 0 % major allele, 100 % minor allele
Having done this, you can be sure that your assay is reliable and can detect differences in allelic expression at least to this level of sensitivity. You can take things further if you want to or need to by changing the ratios of alleles or including more mixes in smaller increments.
Remember Controls
Including a PCR with heterozygous DNA is an invaluable control to determine whether or not your assay detects both alleles, and this control can also help you to normalize your data if necessary.
So, there we have it: several different ways of investigating allele-specific expression using PCR-based approaches. Semi-quantitative methods can tell you whether an allele is expressed at all, and quantitative approaches (which are more expensive) can give you more definitive measures of expression relative to a known standard or relative abundance of each allele.
Measuring allele-specific expression can be an important method of determining, for example, if your gene is silenced during developmental processes or its expression is altered by epigenetic modifications. Alternatively, perhaps your SNP was identified in a genome-wide association study as linked to disease. Allele-specific PCR can help you determine whether that SNP influences gene expression in an allele-specific manner. These questions can be important in cancer, general disease, or in developmental processes.
If you have any comments, suggestions, alternative approaches or improvements, then please leave a comment below.
References
- Gaudet M, Fara AG, Beritognolo I, Sabatti M. (2009) Allele specific PCR in SNP Genotyping. Methods Mol Biol. 578:415–24.
- Abbon, M. Get that Genotyping PCR to Work EVERY TIME. Bitesize Bio.







