Are you stuck in cloning hell?, Tired of doing ligations that don’t work? Want a faster, more efficient cloning procedure? You should try ligation independent cloning.
A growing number of researchers swear by ligation independent cloning methods because they are simpler and more efficient than conventional cloning and as a recent convert to their ranks, I’d like to spread the gospel.
There are in fact a number of ligation independent cloning methods, each of which (as the name suggests) removes the need for ligation, the Achilles heel of standard cloning procedures. In this article I will describe only the ligation independent cloning method that I currently use, T4P-mediated ligation independent cloning or T4p-LIC for short. I’ll deal with the other methods in a later article (article on other methods, article on LIC primer design, LIC protocol).
The principle behind T4P-LIC is simple – instead of using restriction enzymes to generate short sticky ends as in conventional cloning, T4P-LIC uses T4 DNA polymerase to generate long sticky ends of 12-15 nucleotides.
The “stickiness” of restriction enzyme-generated ends is fairly weak the small number of hydrogen bonds between those few exposed nucleotides are not enough to hold the plasmid and insert together permanently, so the covalent joining of the phosphodiester backbone by DNA ligase is needed to stabilize it (see this article for more details). In contrast, the “stickiness” of the long sticky ends generated for T4P-LIC is by itself sufficient to hold the plasmid and insert together, allowing it to be transformed and the backbone repaired by ligases in the host cell.
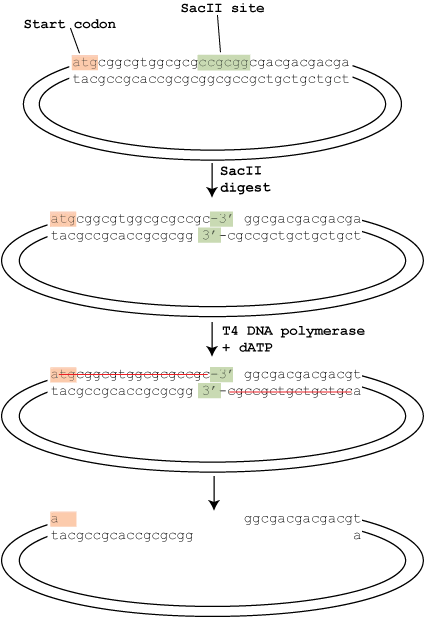
T4 DNA polymerase has two enzymatic activities: a 5′ to 3′ polymerase activity and a 3′ to 5′ exonuclease activity. Given the chance, the exonuclease activity would start at an exposed 3′ end and remove those nucleotides all day. The polymerase activity, however, balances the exonuclease activity by adding nucleotides at the 3′ end. The net result is that in the presence of all 4 dNTPs, nothing happens – every time the exonuclease removes a nucleotide, the polymerase replaces it.
In T4P-LIC this system is cleverly unbalanced to create the sticky ends. As shown in the diagram, the digested vector is treated with T4 DNA polymerase in the presence of only a single nucleotide (dATP in this case). The cloning site of the vector is designed so that going back from the 3′ ends exposed after the restriction digest there are no adenine nucleotides until 18 nucleotides (on the left) and 16 nucleotides (on the right) in. This means that the exonuclease removes the nucleotides, but because dTTP, dCTP or dGTP are unavailable the polymerase can’t balance it’s activity until it reaches the dATP, creating the sticky ends as shown.
The insert is treated in a similar way. The insert is amplified by PCR using oligos with tails that, when treated with T4 polymerase and dTTP, will create overhangs compatible with those on the vector. The vector and insert are then simply annealled together and transformed. Because the left and right overhangs are different, the cloning is directional.
The advantages of T4P-LIC are that the procedure is simpler and more rapid than conventional cloning, the success rate is high – in my experience ligation independent cloning is successful around 90% of the time, and the protocol can be standardized since there is no need to take into account the actual insert sequence (because no restriction enzymes are used on the insert).
Commercial T4-mediated ligation independent cloning vectors are available from Millipore/Novagen and others, but it is very easy to make your own – just design your ligation independent cloning site, have it synthesized on oligos then cut out your current multiple cloning site from your favorite vector and replace it with the ligation independent cloning site. A good example of this can be found here.
If you do a lot of cloning and regularly find yourself in cloning hell, I would definitely recommend that you try out this technique.. your life will be a lot easier!





