Two years ago, all I knew was third BASE in my baseball field and the cutter ball from the pitcher. Now, I know a lot more about lab-based BASES and cutters: REBASE and NEBcutter. While they sound like baseball terms, REBASE and NEBcutter are tools for working with restriction enzymes.
Read on to find out how to use each of these databases.
REBASE’s User Interface Explained
Many of you frequently use REBASE to obtain up-to-date information about restriction enzymes, the workhorses of biotechnology research. The Restriction Enzyme dataBASE (REBASE) was created in the 1970s by New England Biolabs and is patented by Nobel Laureate Sir Richard Roberts. REBASE provides a wealth of information about restriction enzyme type, recognition sequence, and cut site. It also contains information about methyltransferases, crystal structures, and much more.
However, the problem with REBASE is its challenging user interface. When you visit the site, you see an array of image buttons on the home screen. Once you become familiar with the options on this page and understand how to navigate REBASE, you will be able to complete projects much faster with increased efficiency and accuracy.
The REBASE Subdirectories You Should Know
Subdirectory navigation is a considerable challenge with REBASE. Enter a directory and you will notice more image buttons in that directory. For example, click on REBASE sequence data, and you see the nested image button REBASE Tools (red box, Figure 1).

However, these nested image buttons are neither comprehensive nor consistent across all directories.
Never fear. To solve the problem of challenging directory navigation, you only have to learn how to navigate to and between these three directories:
- REBASE Files
- REBASE Enzymes
- REBASE Suppliers
Here’s a quick breakdown of what they do:
REBASE Files gives a lot of raw data, in many different formats, for software programs like BioEdit and SimVector. The data can be uploaded into your genetics software in order to design experiments and analyze results.
Turn to REBASE Enzymes, for an alphabetized list of all restriction enzymes and methyltransferases. Use this option if you want more information on a specific enzyme (but you must know the enzyme’s name).
REBASE Suppliers provides a list of companies that sell enzymes and related products. After navigating through the other directories, you should select a restriction enzyme supplier from here.
A Trick I Learned
Instead of sifting through raw data in REBASE files directory, use the simple search on the home page to search using enzyme name, number, journal name, and organism source. For a detailed search, use the REBASE search directory option. This is very useful if you have specific requirements for your experiment like enzyme molecular weight, temperature, and pH.
NEBcutter
In NEBcutter, you can input linear or circular DNA of up to 200,000 bases. The program returns open reading frames (ORFs), which defaults to the E. coli genetic code, and a list of Type II and III restriction enzymes that cut these ORFs once.
As you can see below (Figure 2), the number and arrangement of options in NEBcutter is confusing to the user, although it is useful.
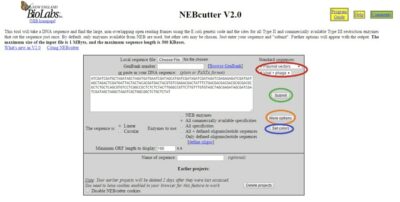
Input
NEBcutter gives you three ways to input data: DNA sequence file, cutting and pasting a portion of sequence, or GenBank number.
At the top right of the grey box (red circle, Figure 2), you can select “Standard Sequences” of plasmid vectors, viruses, and phages. The relevant restriction enzymes for these standard sequences have been pre-identified in the program.
In “More Options” (orange circle, Figure 2), you can select which methylation types to ignore. For example, check boxes for CpG or Dam. You can also change the genetic code to used to search for ORFs in the “More Options” dialog box. The dropdown menu has a variety of options, including mitochondrial genetic code from different species. In the event that you would like to select more enzymes than the default Type II and Type III, you can add enzymes such as nicking enzymes, homing endonucleases, and type I restriction enzymes here as well.
To understand the color format in the output, prior to selecting “Submit”, go to the “Set Colors” option (blue circle, Figure 2) and see the descriptions under the current color scheme.
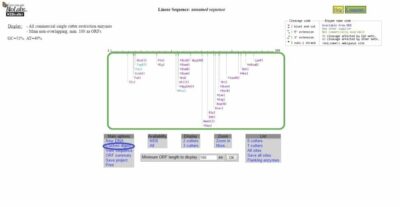
Output
Once you click Submit you will see a map of the DNA sequence with possible restriction enzyme cut sites (green box, Figure 3). If you click Custom Digest (blue circle, Figure 3), there is information on possible restriction enzymes that can digest your sequence. You can click on whichever enzymes you think you might use to digest your DNA.
After this demystification process, you are ready to effectively use REBASE and NEBcutter in your research. Instead of being overwhelmed by the navigational difficulties of these user interfaces, turn your knowledge of the systems to your advantage in your research work.






