The Basic Local Alignment Search Tool (BLAST) algorithm is at the heart of a free suite of online resources available through the National Center for Biotechnology Information (NCBI). While most researchers are aware of BLAST as a sequence alignment tool, NCBI’s BLAST suite offers so much more! I’ll cover in-depth how to use these resources to locate single nucleotide polymorphisms (SNPs) in a gene; design primers with Primer-BLAST; and validate primer targets.
Tip One: How to find SNPs
Given the importance of SNPs in both disease and research, NCBI provides tools for collating a gene’s reported SNPs. To find SNPs, start at NCBI’s homepage and type your gene of interest in the search bar. Select SNP from the All Databases dropdown menu to the left of the search bar, as shown below:
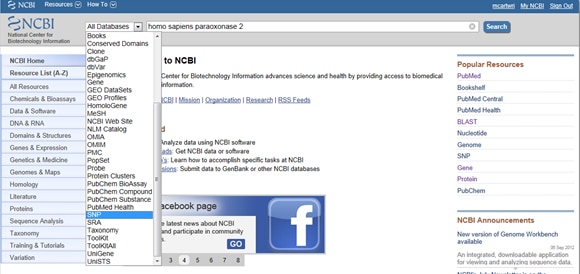
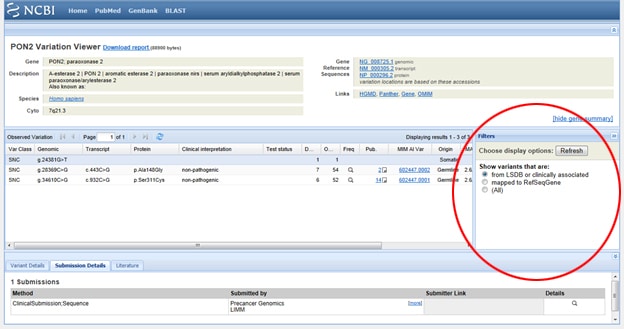
Next, select the light red VarView button below any one of your results:
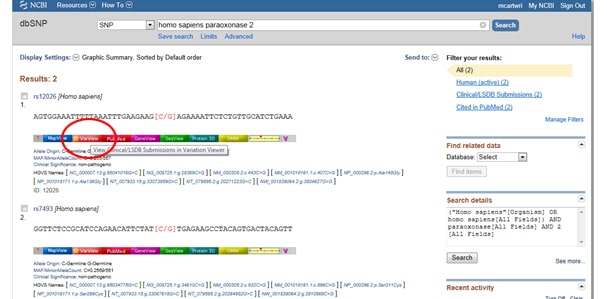
This should bring up reported and annotated SNPs in your query gene, which will come in hand if, for example, you’re designing primers to be used for SNP genotyping or cloning recombinant enzymes.
Trick One: Need to filter the results so that you’re only looking at clinically associated results? Go to that display box labeled Filters to the right of the SNP list under Observed Variation. After you’ve selected your filter option, be sure to hit the Refresh button.
Tip Two: How to Design Primers
NCBI provides Primer-BLAST for automatically designing primers based on a query sequence. To start designing primers, go to the BLAST homepage and scroll down to the Primer-BLAST option under Specialized BLAST. Enter your target sequence either by cut-and-paste or, if it’s listed in NCBI’s databases, as an accession number. I cover some customization options below, but at this point, you can generate primers without doing any additional customization!
Range: At the right of the box for entering your sequence, you can specify the exact range (as numbered 5’ to 3’, from the start of your sequence) of the target that will be considered for designing the forward and reverse primers.
Use my own forward primer (5’->3’ on plus strand): Select this if you’ve already designed your primers and want Primer-BLAST to provide some analytics (e.g. Tm) about them.
PCR product size: Set the range of acceptable lengths of the PCR products here.
# of primers to return: This sets your preferred number of candidate sets of primers to consider. Note that it’s not a guarantee, especially if your parameters are too strict or nonsensical (e.g. you specified a product under PCR product size that can’t be more than 500 bp, but under Range you only want to consider primers more than 1 kb apart).
Primer melting temperatures: This lets you specify your Tm (for a quick refresher on melting temperature, check out our tips for qPCR and regular PCR primer design).
Exon junction span: If you want to exclude genomic DNA (where exons are divided by non-coding introns) then set this to Primer must span an exon-exon junction.
Specificity check: Unless you want Primer-BLAST to return primers that will go off-target (generally not recommended!), leave this checked and specify the Organism your samples are coming from as well as which Database to use, depending on if you’re targeting mRNA, gDNA, etc. By enabling the specificity check, Primer-BLAST will exclude primers that could amplify something outside of your target sequence.
Splice variant handling: If you select this option – only feasible if you’re working off of mRNA sequences – then Primer-BLAST will not exclude primer pairs that could amplify multiple mRNA splice variants of your target. This does not mean, however, that it will give you primer pairs that encompass all known splice variants! You’re simply loosening your target criteria.
Once you’ve entered your sequence and customized as needed, scroll down to the bottom of the page and, after checking Use new graphic view, hit Get Primers. This will return a map of where the suggested primer pairs will amplify your target, as well as analytics on the primers: their length, precise location, respective Tm’s, GC%, and scores reflecting self-complementarity (with 0.00 reflecting no predicted complementation).
Tip Three: How to Predict Primer Targets
How can you check if your primers hit anything off-target? Go to Primer-BLAST. In the query box, enter your forward primer (5’ to 3’). Now type in 20 N’s in a row to separate the primers into individual, non-overlapping alignments. After the N’s, enter your reverse primer (also 5’ to 3’), as shown below:
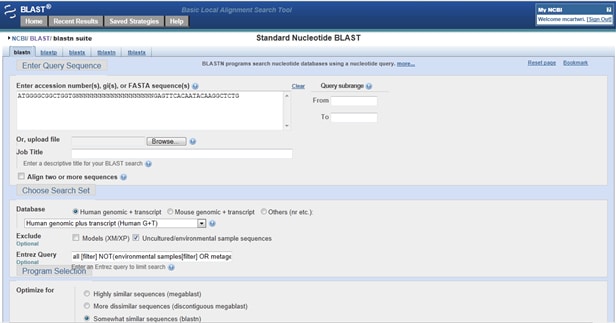
Now you should specify what database(s) you will BLAST your primers against. Are you targeting a human gene and working with human DNA? Then under Database check Human genomic + transcript. Your options aren’t limited to these just human or mouse, however; if working with a different sample – for example, unidentified gut bacteria – select Others (nr, etc.) and pick your database from the dropdown menu immediately below these options (for the gut bacteria example, 16S ribosomal RNA sequences). If you’re working with an individual species outside human or mouse (for example, rat or rabbit), choose among those first three options under Other Databases: Nucleotide collection, Reference RNA sequences (suitable for RT-PCR primers), and Reference genomic sequences.
Once you get your results, check them for certain combinations. If your forward primer aligns on the forward strand (annotated Strand Plus/Plus) and your reverse primer aligns to the same hit, but on the reverse strand (Strand Plus/Minus), then your primers may amplify that hit.
Trick Two: Do your results include things unlikely to have contaminated your PCR samples, such as olive baboons and Neanderthals? If you’re working with human samples or mouse samples, make sure you have those specified under Database. Alternatively, you can exclude specific species.
References and Additional Resources:
Blast Tips. 2007. NCBI. < https://www.ncbi.nlm.nih.gov/feed/rss.cgi?ChanKey=blasttips>
Frequently Asked Questions. NCBI BLAST Help. <https://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=FAQ>
Madden T. The BLAST Sequence Analysis Tool. 2003. <https://www.ncbi.nlm.nih.gov/books/NBK21097/>
Mount DW. Using the Basic Local Alignment Search Tool. 2004. Cold Spring Harbor Protocols. <https://cshprotocols.cshlp.org/content/2007/7/pdb.top17.full>
Wheeler D and Bhagwat M. BLAST QuickStart. 2007. Humana Press Inc. <https://www.ncbi.nlm.nih.gov/books/NBK1734/>