Variability is the Achilles’ heel of research. It can often confound our results and lead us astray searching for solutions. There are two kinds of variability, the first is biological variability. This represents the stochastic nature of the sample you are working with and the inherent differences between samples from the same conditions. There is not much that can be done to reduce biological variation apart from adding more samples to better survey the population and incorporate population-based error (standard error).
The second type of variability is technical, stemming from technique based differences between each replicative sample in your experiment. This type of error incorporates differences in how the samples are analysed by your instrument and slight differences in the performance of your reagents. For quantitative real time polymerase chain reaction (qRT-PCR) there may be several factors that influence technical replicates including slight differences caused by plate variability, or freeze thaw cycles affecting the kinetics of your polymerase, all contributing to variability.
However, the biggest contributor to technical variability is likely you! Or rather your pipetting technique, which may cause some wells to have slightly different amounts of template, polymerase or primers, all of which may be reflected in your data. As qRT-PCR is an exponential reaction, where the region of interest is amplified with each cycle, the effect of slight differences in reagents can often have a cumulative effect on your Ct value. So, what strategies can be employed to reduce the technical variability?
Like a good molecular scout always says: “Be Prepared’
Banish your distractions.
I once worked with a postdoc who refused to let his graduate student do any major experiment that required a lot of pipetting without first eating. If you are hangry (hungry + angry), angry at a reviewer for rejecting your manuscript or distracted by a co-worker who won’t stop singing along with the radio, all of these may factor into your experiment. Try to eliminate distractions so that you have a clear mind when you are pipetting your experiment.
Do the Robot: Have a strategy!
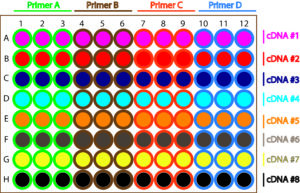
In addition, qRT-PCR tends to have many reactions per experiment, meaning lots of small tubes or wells. If you are not paying close attention, you can easily pipette the wrong solution into the wrong well and may not even notice. I always setup my plates in the same way- my strategy is to load a Master Mix containing my primers and SYBR® mix in replicates in sets of columns, then load my diluted cDNA across the rows (Figure 1).
I always pipette my samples in a certain way: for cDNA, I pipette my control replicates then my experimental ones, with my internal controls (a sample that has not had reverse transcriptase) at the end.
For my primers, I always pipet my housekeeping gene in the first set of columns, then all my primer sets in alphabetical order. Doing it in a set order every time (i.e. alphabetically), means that even if I zone out or become distracted, I can normally work out where I was and I do not even think about the mix I am pipetting, because they are in a pre-determined order.
Bigger is better!
When it comes to pipetting, bigger is always better (within reason!). I make up a master mix which I aliquot into the wells and then I add my diluted cDNA mix (usually 1:20 of my original cDNA). This allows you to pipette a larger volume and be a little more accurate. I normally have a final volume of 20 µl, so my master mix is 11 µl and my diluted cDNA is 9 µl.
Replicate, replicate, replicate
Even if you are a master of the pipette, there will be other differences that you just can’t control. Having two and ideally three technical replicates per sample will enable you to be able to identify if something odd happened in one well so that it can be discounted. This is much more difficult if you have less than 3 wells as you cannot discern which value is the correct one. Although it may use up a little more reagent, I firmly believe that replicates and controls are the key to high quality data. Even if it takes you one more run to analyse all of your samples, you may have saved time and reagents by not needing to re-assay the entire experiment after getting ambiguous results.
Make the reaction work for you
Low levels of your amplicon will result in high Ct values. However, very low levels of your desired product will result in an increased error due to the stochastic nature of PCR amplification. In other words, the less template you have, the more variability you will see in your replicates because the reaction becomes more sensitive to slight changes in amplicon number and template concentration.
To prevent high sample variability affecting their data, many people use a cut-off cycle threshold of 30 when analysing their data, as data points after this may be unreliable. This effect will be apparent regardless of your pipetting skills. To combat this, the easiest thing is to increase you input DNA concentration. Given that PCR results in exponential amplification, a 50% increase in cDNA concentration should push your results by 1-2 Ct values. This is a particularly useful trick when assaying amplicons that have very low expression.
You are only as good as your tools
Keep up to date with lab maintenance
A pipette that is not properly calibrated regularly may dispense more or less than specified. Many companies suggest re-calibrations every 6-12 months and most will come to your workplace to test and re-tune all of your labs pipettes in the span of a day. If you are assaying relative expression and include the required housekeeping gene controls, the variability from your pipette may be fairly constant, hence it may not alter your results a huge amount. However, from a scientific accuracy standpoint, it is a good idea to spend a few dollars to maintain your equipment!
Upgrade your pipetting game
If you have access to multichannel and multi-dispensing pipettes, you should utilize these to streamline and improve your qRT-PCR reactions. In the past, multichannel and dispensing pipettes were associated with decreased accuracy, however they have been improved greatly with time and are now often more accurate than single-dispensing, single channel pipettors. To dispense my master mix I use a single channel multi dispensing pipette. For my cDNA, I dilute it into 8-strip PCR tubes and use an 8-channel pipette to dispense the cDNA into my reaction wells (Figure 2).
Understand your reagents.
SYBR-Green® and many qRT-PCR master mixes often contain detergents such as glycerol which make the solution vicious. Thus, you should not pipette these viscous solutions as you would water, as it is much thicker and doing so will likely result in you under-pipetting. The best practise is to aliquot your master mix to use reverse pipetting. This will pre-wet your pipette tip and help to pipette more accurately.
Hopefully with these tips you can up your pipetting game and generate more consistent and better quality data! Keep calm and pipette on!








