If you’ve ever performed PCR, you’re probably already very familiar with DNA oligonucleotides (or oligos). But did you know that these molecules can do so much more than just act as simple primers? You can add a wide range of modifications to your oligos, which may change the stability, binding, solubility and even visibility, to expand their scientific and therapeutic uses. Here, we will introduce just a few of the available modifications.
Phosphate
Figure 1: 5’ and 3’ phosphate modifications.
There are two common reasons you might wish to order a phosphate-modified oligo: to ligate multiple sequences together or to block polymerase extension in a qPCR reaction. Ligation reactions typically require phosphorylation of the oligonucleotide terminus allowing the construction of a longer sequence from multiple oligos or PCR products.
Addition of a phosphate group on the 3’ end is also handy if you need to block elongation of a dual-labeled probe during your qPCR reaction when the quencher is placed internal to the sequence. Although you should note there are other modifications, such as a C3 spacer, that provide a more robust block.
Carbon Spacers
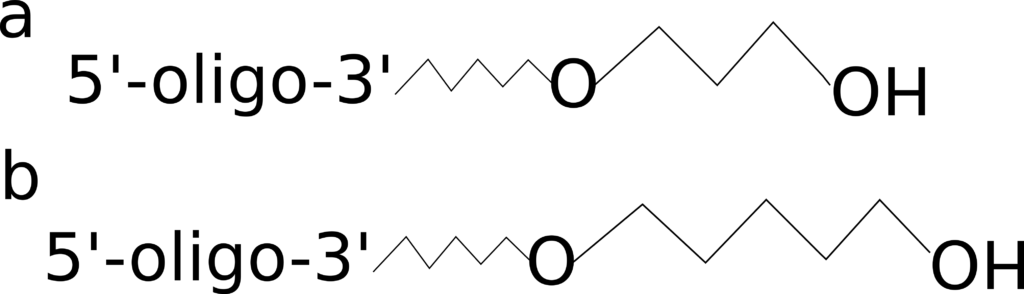
Figure 2: 3’ C3 and C6 spacers.
In situations where you are adding a modification to your oligo, you may also require inclusion of a spacer, which come in a variety of sizes, including C3 and C6, shown in Figure 2. In cases where the proximity between the attached modification and the oligo may affect activity, e.g. due to steric hindrance, multiple spacers can be combined to create longer ones. Companies will often add spacers for you, depending on your needs. Spacers can be used to change the hydrophobicity of the attached modification or to increase flexibility of the oligo, since this modification disrupts the sugar phosphate backbone. The addition of a spacer to the 3’ end can also block both polymerase extension and ligation.
While the above rings true for carbon spacers, there are a wide range of different spacers that can be used, each having different effects on the various properties of your oligo.
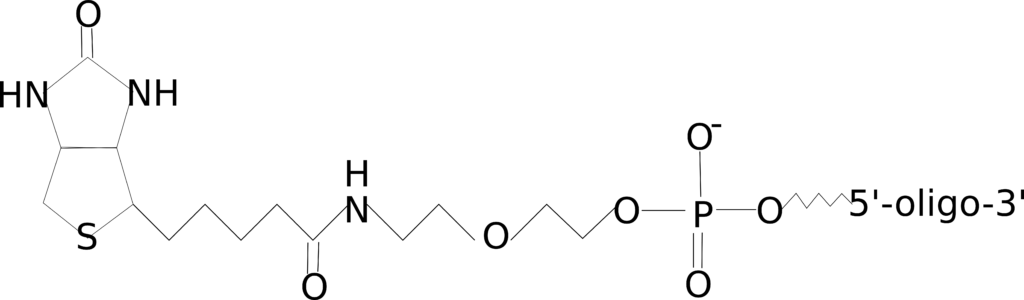
Biotin
Figure 3: 5’ biotin modification.
Biotin, as you may already know from the article on biotin-streptavidin for immunohistochemistry, specifically binds to avidin. This biotin-avidin interaction can be exploited to detect your oligo or capture or purify it from a complex mixture. There are a number of methods that utilize this biotin-avidin interaction, such as Chromatin isolation by RNA purification (ChIRP), which is used to investigate the binding location of non-coding RNAs, such as lncRNAs and enhancers.
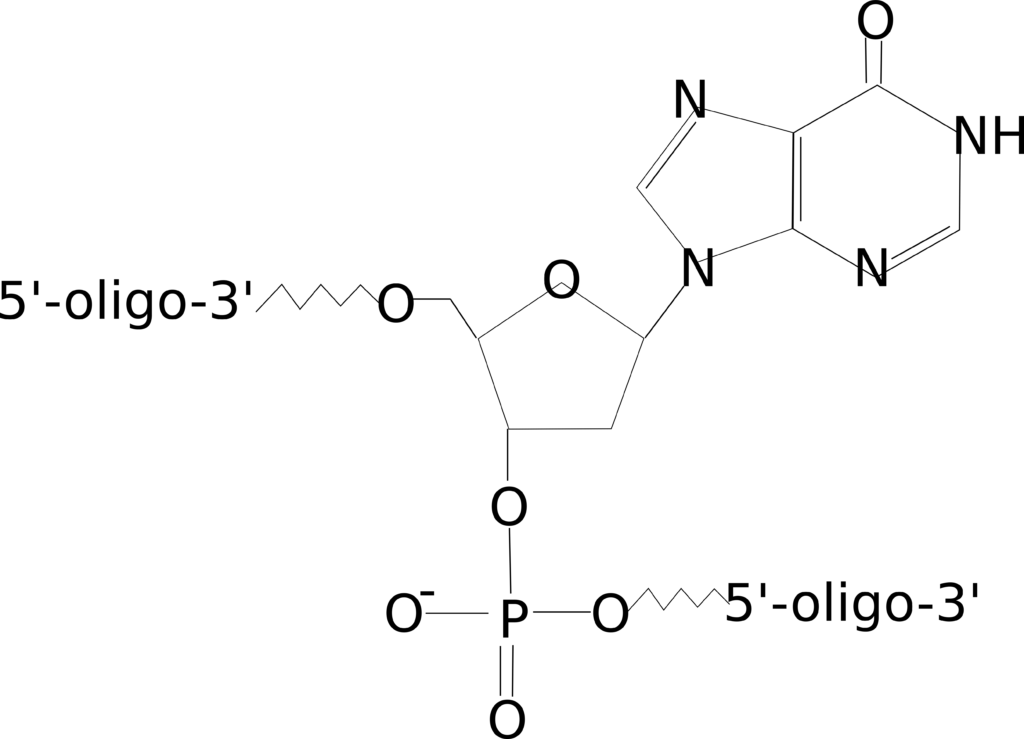
Wobbles and Inosine
Figure 4: Internal inosine modification.
Do your samples have SNPs? Or are you trying to detect multiple variants of a particular target (such as looking at several strains of a microbe)? If so, degenerate (ambiguous) bases can be replaced with a wobble, or mixed base. The incorporation of wobbles into your oligo design will generate a mixture of oligo species that differ at that specific site in order to represent each sequence.
You can choose to add a two-, three- or four-base wobble, depending on the variation of bases that appear at that location. However, if you are unsure which base occurs, or if all four occur, it may be preferable to generate one oligo species containing an inosine, which binds to all four DNA bases, at that particular site.
A note of caution when working with wobbles – when calculating the amount to use in a reaction you should consider the concentration of the individual species rather than the total concentration of oligos. For example, if you add a single two-base wobble the effective concentration of the oligo is halved, and you should therefore correspondingly increase the amount of oligo in the reaction to prevent any particular species from limiting the reaction kinetics. For this reason, wobble bases should be used sparingly in a design.
Phosphorothioate
This is a modification not of the bases themselves, but of the linkage between bases. The incorporation of phosphorothioate linkages (as opposed to standard phosphodiester linkages) between bases protects oligos from the action of nucleases. Thus, this modification is particularly useful for antisense oligos that you will be transfecting into a cell. The number and placement of these modifications required to protect against nuclease activity isn’t certain, so a bit of trial and error may be necessary to achieve the desired level of protection. You can learn more about phosphorothioate bonds in The BiosearchTech Blog.
2’-OMe: The “2-O-Methyl” bases
These modified RNA bases are another way to protect your oligos from nuclease activity. 2-O-Methyl bases do have their downsides, as they aren’t recognized by many enzymes, potentially limiting their uses. However, this modification can be combined with the above phosphorothioate modification to produce oligos that are very resistant to nuclease activity.
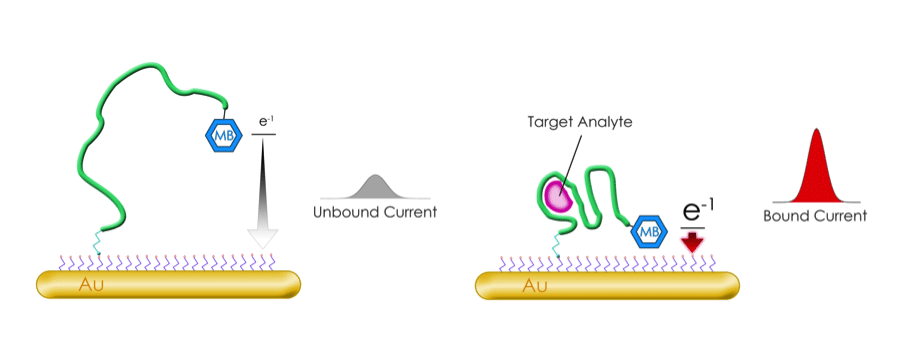
Methylene Blue
Figure 5. Methylene blue modification converts oligos into electrochemical biosensors. Image reproduced with permission from Biosearch Technologies.
This is a pretty cool modification which, when added to an oligo, allows it to act as an electrochemical biosensor. In short, when conjugated to an electrode, the methylene blue modification allows the detection of conformational changes in the oligo. As methylene blue moves closer to or further away from the electrode, the transfer of electrons between the two is altered, indicating binding of the oligo to the desired target. Monitoring the electron transfer between methylene blue and the electrode provides a specific and sensitive way to detect the target in a sample. You can read more about methylene blue in the The BiosearchTech Blog.
A Multitude of Modifications
This article covers only a snapshot of the different modifications available for your oligos and the variety of applications they have. So if there is something you need your DNA to do – light up, bind, stay put or chemically react – take the time to check out the different modifications available before ordering your next oligo.
If you want to find out more about the different modifications and what they do, keep an eye on the The BiosearchTech Blog, where we will be highlighting other modifications and their myriad of applications.