Understanding the basic (and simple!) chemistry behind the most commonly used molecular biology techniques in the lab won’t just enhance your understanding of the science; it will help you get better results. Read on to get a short primer on the basics of DNA ligation, along with some handy hints on optimizing your DNA ligation reactions.
The Enzyme Doing the Work: DNA Ligase
DNA ligase (EC 6.5.1.1) is the enzyme at the heart of the DNA ligation reaction. Simply put, DNA ligase sticks two bits of DNA together! Or, to be more scientific about it, DNA ligase covalently joins the phosphate backbone of DNA with blunt or compatible cohesive ends (see Figure 1).

In living organisms, DNA ligases are important enzymes with key roles in DNA replication and repair, specifically in repairing double-strand breaks in DNA molecules.
DNA ligation is performed for both cloning and non-cloning applications in the lab. One example of the uses of DNA ligation for non-cloning applications includes the construction of next-generation DNA sequencing libraries. These processes use DNA ligase to connect specialized adapters to both ends of DNA fragments to allow for subsequent sequencing.
For cloning applications, DNA ligase is commonly used for the insertion of restriction enzyme-generated DNA fragments into vector backbones. Commercial ligases are supplied with a reaction buffer containing ATP and Mg2+, both of which are essential for ligase activity.
The Two Steps of the DNA Ligation Reaction
The DNA ligation reaction itself has two basic steps:
- First, the DNA ends have to collide by chance and stay together long enough for the ligase to join them.
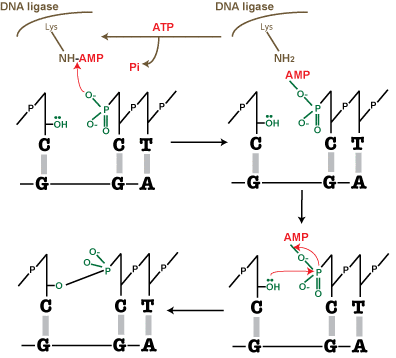
- The second step is the enzymatic reaction, which is shown schematically in Figure 2. DNA ligase catalyzes the joining of the 3′-OH to the 5′-phosphate via a two-step mechanism.
First, the AMP nucleotide, which is attached to a lysine residue in the enzyme’s active site, is transferred to the 5′-phosphate.
Then the AMP—phosphate bond is attacked by the 3′-OH, forming the covalent bond and releasing AMP. To allow the enzyme to carry out further reactions, ATP must replenish the AMP in the enzyme’s active site.
6 Tips For Optimizing DNA Ligation Reactions for Cloning
DNA ligations can be frustrating. Sometimes they fail to work for no obvious reason.
Our top 6 DNA ligation tips should improve the efficiency of your ligations and will hopefully increase your cloning success rate!
1. Lower the Temperature
The second part of the DNA ligation reaction is the most inefficient part of the reaction, but it is easier at low temperatures.
Why? Well, as you will probably know, all molecules move faster at higher temperatures, so you can imagine that it is going to be easier for two DNA ends to collide and stay together if they are gently floating through the solution at low temperature rather than whizzing about as they would be at higher temperatures.
There is an additional reason for cohesive ends. Lower temperatures stabilize the hydrogen bonding between the complementary nucleotides, which really helps to keep things in place.
The DNA ligase enzyme has optimal activity at 25°C, so the ligation reaction is carried out at a temperature that is a trade-off between the optimal temperatures for bringing the DNA ends together (1°C) and the enzymatic reaction (25°C).
Normally 1 hr at 16°C is fine but since bringing the DNA ends together is the least efficient part of the reaction favoring this by lowering the temperature to 4°C can give even greater efficiency. However, the enzyme will work very slowly at this temperature, so a long (e.g., overnight) incubation time is required.
2. Aliquot the Ligase Buffer
The ATP in the ligase buffer is essential for the DNA ligation reaction but can be broken down by repeated freeze-thaw cycles. To avoid this, aliquot the ligase buffer from each new stock of DNA ligase.
Make the aliquots small enough for single-use (e.g., 5 µL), and make sure to completely defrost and mix well before you aliquot.
3. Heat the DNA Just Before Ligation
When setting up a cohesive-ended ligation, mix the vector and insert fragments first, and heat to 65 °C for 5 min before adding the remaining reaction components.
This heating step disrupts any vector/vector or insert/insert cohesive-end interactions that may otherwise interfere with the desired vector/insert interaction, reducing ligation efficiency.
4. Check the pH
The optimum pH range for DNA ligation is between 7.6 and 8.0. Depending on how the DNA fragments were prepared, the pH of your ligation mixture may lie outside of this range.
You can check the pH of your ligation mixture by pipetting approximately 0.2 µL of the mix onto narrow range pH paper (e.g. pH 6-8). If required, adjust the pH using 0.2 µL drops of 2M Tris base or 1M HCl.
5. Include Polyethylene Glycol (PEG)
As with any chemical reaction, the concentration of the reaction components can greatly influence the speed of the ligation reaction. PEG is a hydrophobic molecule that takes up space in the reaction, effectively increasing the concentration of the aqueous reaction components, for example, DNA, ATP and ligase.
Adding PEG (e.g. PEG 8000) to a 5-15% final concentration may increase ligation efficiency. Bear in mind that PEG concentrations above 5% can reduce transformation efficiency. In addition, heat inactivation or extended incubation of ligation reactions containing PEG can also decrease transformation efficiency.
6. Add a Restriction Enzyme Just Before Transformation
This neat trick can be used to circumvent high background resulting from undigested vector. If the vector fragment removed during the preparative digest contains a unique restriction site, adding the respective restriction enzyme to the ligation reaction will selectively digest any intact vector, preventing it from being transformed.
Adding 1 µL of the enzyme 5-10 minutes prior to transformation should be sufficient.
DNA Ligation summarized
Hopefully, we’ve shown how optimal conditions for DNA ligation reactions are a delicate balance between DNA molecules interacting and the enzymatic ligation reaction. We hope this information and our top tips help you achieve ligation success.
Want to know more about DNA ligation? Discover why T4 DNA ligase is such a popular ligase and perhaps the only ligation enzyme you’ll ever need.
Need a helping hand calculating the amount of insert for your ligations? Download our free lab math cheat sheet for a handy formula and lots of helpful calculations.
Have we missed a top tip for ligation reactions? Leave a comment below!
Originally published on 31 October 2007; updated and republished May 2022.





