Whether cloning is your everyday work, or you just dabble, you are bound to meet the situation where you need to remove a stretch of unwanted DNA from your vector, e.g., removing unwanted DNA sequences or making an insertion.
If you are lucky, two rare-cutter restriction sites will neatly flank the piece you want to remove, allowing you to easily drop out without any problems. But rarely does Murphy’s Law make anything so easy.
But never fear. Should you find yourself in this situation without a unique restriction site to be seen, we have two alternatives that can break the laws and set you free on your way to your cloning goal.
This article was inspired by a question from one of our readers (thanks, Derek!) who asked us to provide some tips for the removal of a small piece of DNA (60 bp) in a multiple cloning site of a larger vector (8 kb) to prepare it for cloning a new gene.
Tips for Removing Unwanted DNA from Vectors
Exonuclease Digestion
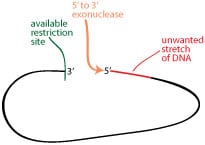
The first alternative to restriction sites for removing an unwanted piece of vector is exonuclease digestion. This could work for you if you have an available restriction site on at least one side of the DNA you want to excise, and if you don’t have to be completely exact about the amount of DNA you remove.
Exonucleases chew away one end of a single-stranded piece of the dsDNA in a controlled and linear manner. So after digesting with your available restriction site, you can use an exonuclease to chew away the unwanted stretch of DNA. There are a variety of exonucleases available, so be sure to pick one that does what you need. Of course, exonucleases can chew the DNA in either 5′ or 3′ direction, so be sure to choose one that moves in the direction you need. The schematic example on the right shows the use of a 5′-3′ exonuclease.
To control the amount of DNA removed, perform a time course experiment with linear DNA and look at the change in size over time to find the amount of time that removes only 60 bp. A high percentage agarose gel should allow you to analyze small changes in size with enough accuracy to make an estimate.
This protocol does not allow for exact knowledge of how much DNA was removed, but depending on what you need, getting close to the target can be good enough.
Exonucleases are pretty useful things—they are also central to ligation-independent cloning, which we covered in another article.
PCR Amplification
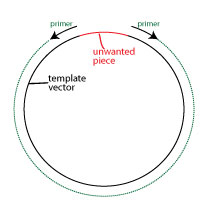
The second alternative is more exact. It involves performing a PCR reaction to amplify the vector using primers that are in opposite directions to each other, and which leave out the unwanted region.
This requires a high fidelity enzyme that can amplify long products (out to 8kb in Dave’s example). One example is the Phusion enzyme by Finnzymes. This enzyme can amplify up to 20 kb with double the fidelity of standard proofreading enzymes. Another option is the Thermo Extensor enzyme blend. There are several options for long PCR with high fidelity, so if you have a favorite supplier, I recommend you check them out and see what they offer.
By using PCR primers amplifying in opposite directions, you will generate products that lack the unwanted DNA. You can use an exonuclease for ssDNA to remove any overhangs and blunt end the vector.
A second round of PCR (this time using a non-proofreading polymerase like Taq) can be used to add back “A” overhangs if you want to clone into that space another PCR product. Or, the blunt-ended DNA can be re-ligated together to circularize the plasmid so it can be transformed back into E.coli for preparing a larger batch of your new vector.
A Final Word on Murphy and Plasmids…
Removing unwanted DNA from vectors is one of the most routine laboratory techniques in cloning, and so is ripe for the application of Murphy’s Law, and can be the problem of much frustration.
When the typical approaches don’t work, sometimes a completely different approach can help you beat Murphy’s law. Hopefully, some of these ideas will help you do so, and even inspire you to further creative ideas to get the vector you need.
Incidentally, if you want to know a bit more about the science of Murphy’s law, take a look at Matthews, R A J (1995). “Tumbling toast, Murphy’s Law and the Fundamental Constants”, a 1995 European Journal of Physics (and Ignoble prize-winning) article by Robert Matthews, which explains why toast always falls buttered side down.
If you are having problems with a particular aspect of your lab work, please feel free to leave any comments below. We can’t guarantee we’ll answer all your questions, but if yours catches our eye, we’ll write our suggestions up in an article just like this one.