Microarrays are one of the most in-depth ways of determining cellular gene expression levels of thousands of genes simultaneously. They are able to help determine:
- Gene function and cellular processes
- Gene regulation and interactions
- Gene expression levels in different cell types and how this expression is altered by the addition of various compounds or disease treatments
Getting good microarray results starts way before analyzing your data, however. A crucial first step in setting up a good microarray experiment is extracting high-quality RNA from your cells. In this article, I’ll discuss the various options you have for extracting RNA, how to handle it, and how to analyze its quality.
RNA Extraction
If you look at the literature, people often prefer to make up their own solutions and use their own protocols rather than use an RNA extraction kit. This is all down to personal preference and what you want to do with your RNA at the end of the extraction process. Personally I found RNA extraction kits to be fine for this kind of work.
Most companies provide RNA extraction kits; however, some are regarded more highly than others. A good website to use to compare scientific products from a wide range of companies is www.biocompare.com. This will allow you to look at kits provided from different companies and determine which one is best for you. All of these kits will come with their own specific instructions as to how to extract RNA from your cells. Alternatively, you can make up your own solutions; however, if you are new to the process of RNA extraction this may be more complex.
Whether you choose to use a kit or make your own solutions, the basic steps of RNA extraction are as follows:
- First, guanidium thiocyanate is added to the cells along with a reducing agent, such as beta mercaptoethanol, which will disrupt cells and break disulphide bonds as well as acting to inactivate proteins (including those pesky RNases!). After adding these components, the sample is normally subjected to vigorous shaking or vortexing.
- Next is phenol-chloroform extraction. Chloroform dissolves lipids and phenol solubilises any proteins. After addition of phenol and chloroform-isoamyl alcohol and sufficient mixing, two phases will form in the tube. It is the clear upper aqueous phase that will contain your precious RNA.
- After removal of the RNA-containing phase to a fresh tube, isopropanol can be added to precipitate it. After centrifugation, the pellet is then washed with 75% ethanol to remove any impurities.
- The pellet can then be resuspended in DNase- and RNase-free water and stored at -80oC
The most important thing to remember here is that RNA is extremely sensitive! The slightest speck of RNase in your sample will leave you with poor quality RNA, which is not good news! It is essential that EVERYTHING you use is sterile. Clean everything with 100% ethanol or if you want to be doubly sure, then you can buy DNase- and RNase-away from commercial suppliers.
Checking the Purity of the RNA
The best way to check the purity of your extracted RNA is to run a sample on a BioAnalyzer (you’ll probably have to send an aliquot of your samples somewhere else to do this). You will then receive an output like the one shown below:
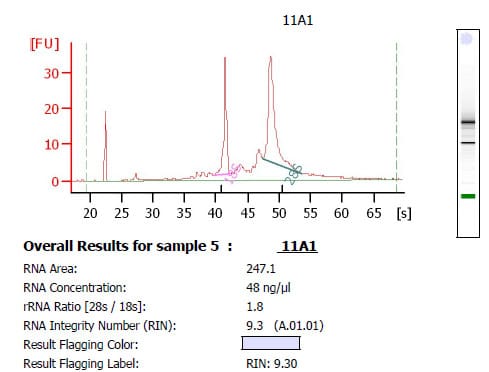
The three things that you need to check are:
- That you have two clear peaks: one for the 18s and one for the 28s ribosomal subunits
- That the ratio of the two peaks is between 1.8 and 2.1
- That the RNA integrity number (RIN) is as close to 10 as possible
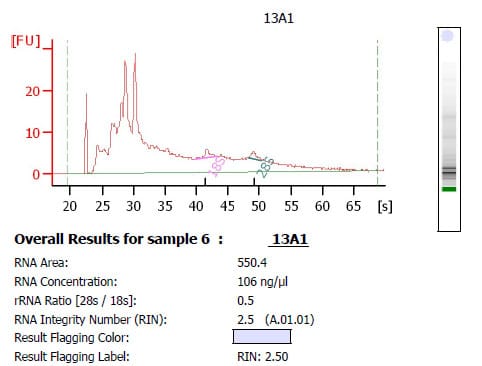
If your results meet all three conditions listed above, then congratulations, you have RNA of sufficient quality to be run on a gene expression chip! However, if your RNA outputs look more like the one below then you may have a bit more work to do first.
The output above demonstrates a poor result from the BioAnalyzer. The 18s and 28s peaks are unclear and the majority of the peaks are on the left-hand side of the graph. This indicates degradation, which is verified by an RIN value of 2.5, which is not really useable for gene expression studies. The ratio of the 18s:28s peaks still falls within the required region of 1.8-2.1; however, this does not mean that the RNA is suddenly ok to use, as the BioAnalyzer output must meet all 3 of the conditions listed above!
If you end up with poor quality RNA, then you should think back to how you prepared the RNA. It may be because things were not clean enough or that those cheeky RNases got involved somehow! It may also have been that the RNA had been freeze/thawed too many times as this can damage it greatly. Ideally you should aim to freeze/thaw your RNA samples only once. Unfortunately, the bad news is that you will have to re-extract your RNA from your cells and try again.
What are your tips for successful RNA extraction?





