What if you could see intact structures inside cells, in three dimensions, with the image volume exceeding tens of microns, at nanometer resolution?
All of those are possible with volume electron microscopy (vEM), a dazzlingly powerful offshoot of electron microscopy (EM).
Read on for an introduction to vEM, how it works, the questions it can address, and some of its biggest challenges.
What Is Volume Electron Microscopy?
Simply put, vEM enables us to image three-dimensional (3D) structures of biological samples at high resolution in their native environment. The term describes a range of electron microscopy techniques that provide a 3D image at the end rather than a single linear method.
Such a summary belies its power, however. There’s a reason Nature listed it among their technologies to watch in 2023, along with titans like the James–Webb telescope and high-precision radiocarbon dating.
3D images of large chunks of cells and tissues allow us to see and interrogate networks of subcellular components. Generally speaking, the more connected subcellular components (such as mitochondria and synapses) are, the healthier the cells are. Plus, organelles and molecular complexes associate and dissociate with each other and their environment according to their function.
The reverse is also likely to be true. Disconnection between subcellular components might indicate pathological states for a given sample type—and this is an area vEM lets us explore!
Besides connectedness, there are other benefits of vEM too. You can analyze a much bigger area than conventional EM methods but without compromising on the resolution of your data or processing additional samples. The increased imaging area means you can image multiple cells simultaneously and choose larger regions of interest.
We’ll come to the research questions you can answer using vEM later, but for now, note vEM reveals sample connectedness, texture, and spatial/cellular context.
The key word here is connectedness.
vEM originated within connectomics—studying the brain’s intact structural and functional connections called the connectome—which is challenging to study with traditional 2D imaging methods.
Fluorescence microscopy accurately captures relatively macroscopic properties, such as the location of cellular components, while electron microscopy captures the ultrastructural details of a sample. However, both these techniques are usually limited to the horizontal plane.
This explanation is slightly simplified—we have confocal laser scanning microscopy, after all. But that requires getting your target to fluoresce by conjugating it to dye or using transgenics. The point is vEM is an additional 3D imaging technique that has its own key benefits.
Why Do We Need vEM? A Simple Illustration
So why do we need vEM? Unfortunately, 2D slices and cross-sections through 3D shapes can lead to misleading representations of their shape.
Check out Figure 1 below to see how many cross-sections a cube can generate—it depends on how you slice it!
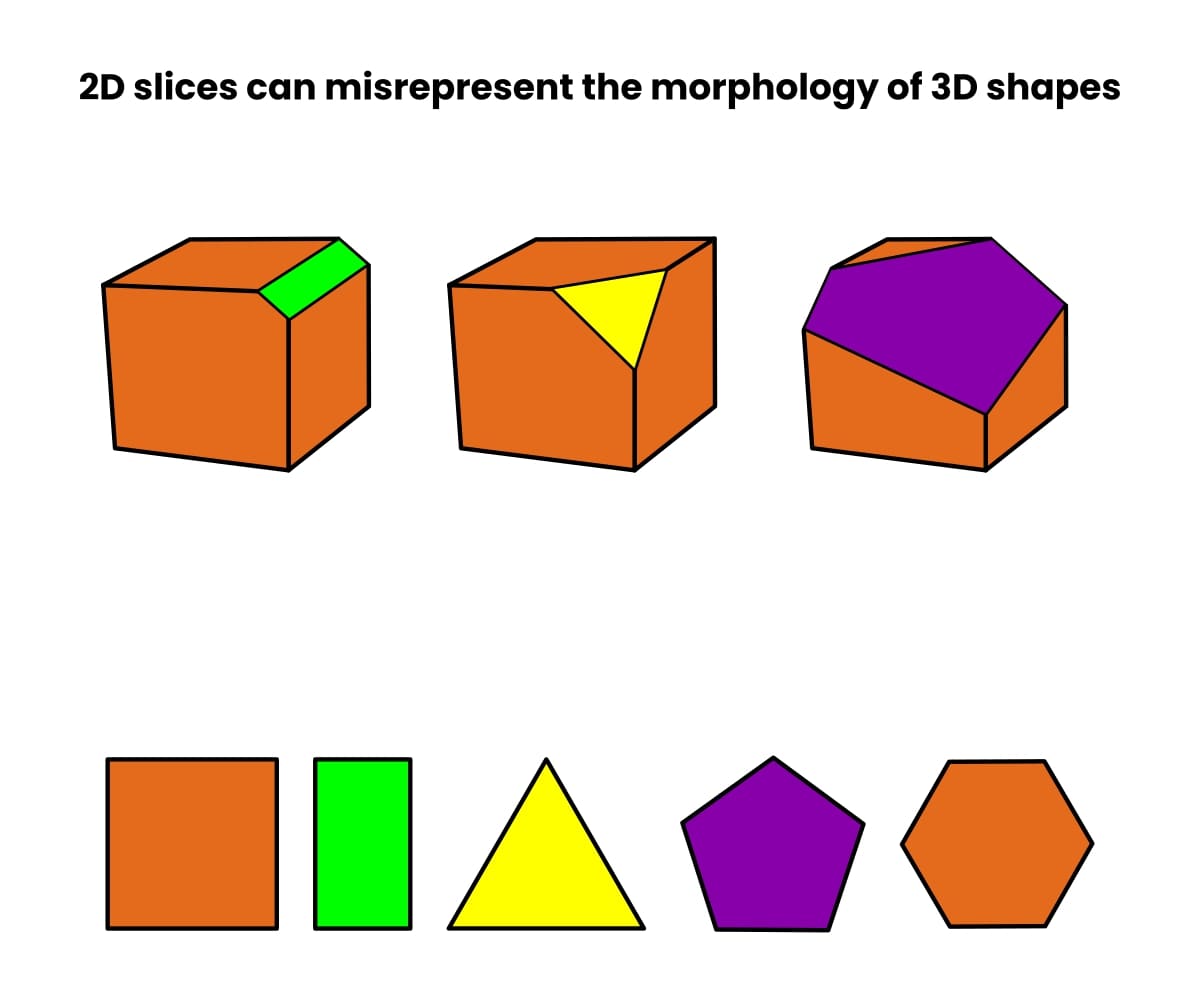
Now imagine that, rather than a cube, you have an organelle such as a vacuole or mitochondrion. The number of possible (and misleading) cross-sections is almost limitless.
Then imagine that you are looking at a structure that no one has ever seen. How do you know what’s accurate?
One solution is to get a 3D image of the intact structure. And now, with vEM, you can!
How Does vEM Work?
Conventional electron microscopy methods usually involve sectioning samples into single, ultra-thin slices and imaging them.
With vEM, you acquire images from a series of slices through your sample and then computationally combine them to create the final 3D reconstruction. And what a reconstruction! Check out this 3D rendering of several connected neurites. [1]
There are three main ways to generate multiple slices from a sample for 3D imaging.
1. Array Tomography
With Array tomography, you fix and section your sample as you would for any other electron microscopy approach, but you generate a lot of sections. Then, you arrange them into a ribbon that serves as a ticker tape through your sample.
Once you’ve got all your sections, you image them serially to create a 3D reconstruction.
Plus, it’s non-destructive, meaning you can retrieve the sections and image them using any other modality. It’s also compatible with scanning electron microscopy (SEM) and transmission electron microscopy (TEM) so long as you prepare correspondingly suitable sections. Read this article for a quick refresher on the differences between SEM and TEM if you need it, and check out this article for a deeper dive into array tomography.
2. Serial Block-Face Scanning Electron Microscopy
The serial block-face (SBF) approach is similar to array tomography. However, instead of generating all the sections at the start and imaging them all in series, the sectioning and imaging are iterative. That’s to say, you section, then image, section, image, and so on.
The great thing about SBF is that you can automate it. The Volumescope 2 SEM is a powerful, modular vEM solution that contains an ultramicrotome inside the imaging chamber for automated data acquisition!
However, unlike array tomography, SBF is destructive, meaning analysis via other microscopy methods must come beforehand. Plus, it’s only compatible with SEM at the time of writing.
3. Focused Ion Beam Scanning Electron Microscopy
Focused ion beam scanning electron microscopy (FIB-SEM) combines a focused ion beam (usually gallium ions) with scanning electron microscopy. Unlike serial block-face imaging, FIB-SEM does not involve removing individual sections using a microtome. [2]
Instead, it uses the focused ion beam to erase sample material layer by layer to reveal a new, deeper surface for imaging at each step. Because of this, it’s destructive.
On the plus side, using an ion beam to remove sample material affords a much higher resolution in the z direction when compared to using an ultramicrotome. The z-direction resolution can be as high as a few nanometers.
Furthermore, the Hydra Bio Plasma-FIB goes beyond gallium ions and features xenon, argon, oxygen, and nitrogen ion plasma sources. The choice of ions allows you to match the ion to the sample or use one ion to (say) quickly excavate your region of interest and another to polish its surface. Watch Cryo-tomography to Volume EM: Explore with Hydra Bio Plasma-FIB to learn more.
How to Decide Whether to Use vEM
vEM is amazing. But should you use it?
The best thing to do is sit down with your local analyst or core facility manager and speak to them about the goals of your project to decide if it is appropriate.
vEM takes a long time (see below), and the facilities to do it are rare at present, so you must formulate your research question well and have clear, definite goals.
There are two essential criteria:
- Your research question must be concerned with 3D ultrastructures.
- This ultrastructure must confer a change in function or phenotype.
Here are some examples of questions you can address using vEM:
- Are there changes in cellular/organellar shape/organization under different conditions?
- Are there changes in cellular/organellar connectedness under different conditions?
- Are there ultrastructural differences in tissues or cells under different conditions?
- Where do subcellular entities originate, how are they distributed, and where do they go?
And, because vEM enables you to locate pretty much any subcellular component you want (providing you can stain it), you can start to answer deep questions such as:
- How much of X is there per cell?
- What is the total volume and mass of X per cell?
- How do these values change under different conditions?
Perhaps the key point is obvious—the raw data you answer these questions with is an image, not an abstract representation of your data.
And vEM datasets are incredibly rich. Plan for this and prepare to take the time to analyze your data carefully. Bear in mind that there will probably be more data than you can hope to interpret, and your images will likely contain structures invaluable to someone else’s project. Sharing your data is therefore encouraged.
The Electron Microscopy Public Image Archive is the primary online repository for 3D images and tomograms. Be sure to look at some of the incredible data on it.
The vEM Workflow
So, you have decided to take the plunge and embark on a vEM project. What can you expect?
Before you do anything, visit the volume electron microscopy community’s webpage. It hosts training videos, protocols, explanations, key people, news, a Twitter feed—everything!
Here is a breakdown of the workflow, key considerations, and things you can expect. If you’re in a hurry, check out Figure 2 below.
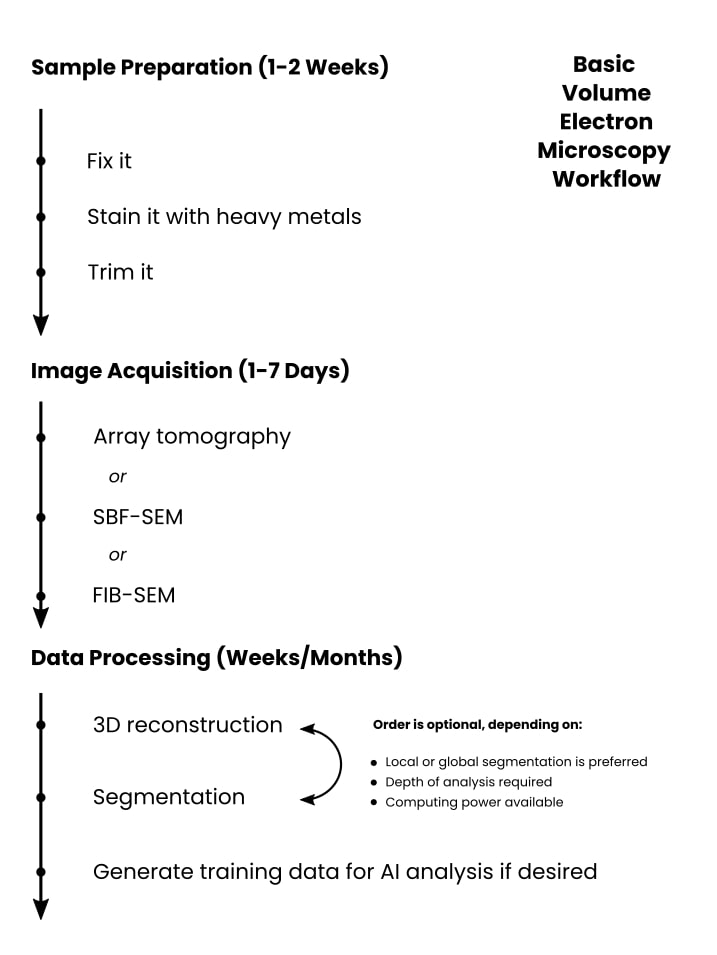
Sample Preparation (1-2 Weeks)
Sample prep for vEM has a lot in common with other EM techniques. You fix your sample, coat it with a heavy metal stain to provide contrast, then slice it up and image it.
You’ll want to identify your region of interest using a sensible approach such as fluorescence microscopy or micro-computer tomography (micro-CT). Then you’ll move on to staining your sample before embedding it in resin and doing any manual trimming.
Take care with your sample prep, as it largely dictates whether your vEM imaging project will succeed or fail. We have an excellent guide to EM sample prep, and many of the points apply to vEM also.
And finally, note that vEM samples are often embedded in a non-conductive resin, so one of the challenges is ensuring that your sample prep results in conductive samples for imaging.
Image acquisition (1-7 Days)
As mentioned earlier, a key technology in vEM is SBF imaging, where your sample is cut into sections using an ultramicrotome inside the imaging chamber of the electron microscope.
In such cases, image acquisition starts by carefully aligning the ultramicrotome to work harmoniously with the microscope optics. Then, you set the cutting window and imaging parameters before acquiring your data.
Currently, vEM instruments are large and expensive. The data acquisition does not tolerate sample movement, so they are usually on anti-vibration floors and need shielding from electromagnetic interference from surrounding infrastructure (train stations, trams, etc.)
Data Processing (Days, Weeks, or Months)
Once you have data, you need to process and interpret it. How long this and your hands-on time will take depends on your research goals.
One step you’ll have to do, regardless of your downstream intentions, is to align your slices to produce the 3D image.
Beyond that, it depends, as I say, on your goals. In theory, inspecting your final 3D image for a yes/no answer to your research question should be relatively quick and simple.
But you may need to do something more complex, such as identify regions of interest to segment to (say) generate training data for an artificial intelligence model. Depending on your sample type, it may be hard to segment using automated methods, meaning you must do it manually. If you’ve generated a lot of data, this could take years!
However, most projects won’t be at this extreme edge of things, and weeks to months is probably a more reasonable estimate.
And remember, vEM technology is in its infancy. Technological progress, driven by ambitious research and data-sharing initiatives, will doubtless reduce these time frames to something more feasible.
The Current Challenges of vEM
So why isn’t everybody doing it? vEM and the community surrounding it are relatively new. Several challenges prevent the technique from being mainstream (for now).
Apart from sample preparation, which is user-dependent, some of the challenges are as follows.
Data Collection Time
vEM projects are intrinsically large-scale, and the data acquisition speed of electron microscopes limits the amount of data collected in a given period. And unsurprisingly, there’s a trade-off between data acquisition speed and data resolution.
Insufficient Hardware Automation
And speaking of large-scale projects, electron microscopes of any kind are complicated. Set up and monitoring require dedicated personnel, meaning the average user can’t just queue up a load of experiments and walk away. The emergence of vEM may push electron microscope manufacturers and facilities toward greater automation.
Handling Vast Quantities of Data
Data handling and analysis are also limiting steps because vEM projects can generate terabytes of data. You can’t just slap it on a memory stick and take it to the office. Such vast amounts of data are challenging to transfer, let alone analyze and interpret. And you’ve got to back it all up!
Data Segmentation
Imaging larger areas has two major consequences: there’s more data to segment, and the data are more biologically complex. Imagine segmenting all the mitochondria in a cell—you get the idea. Fortunately, machine- and deep-learning methods are constantly evolving, meaning that we are always heading towards more accurate, automated segmentation.
If you genuinely have to segment a lot of mitochondria, check out the AI tool MitoNet. [3]
Insufficient Data Analysis Automation and Streamlining
Similarly, automatic image annotation and segmentation methods are still insufficient to annotate such rich and vast datasets accurately and reliably. Double-checking the results of automated annotation also takes time. Assuredly data analysis and software pipelines will develop tremendously.
3D Reconstruction is Hard
Then there’s the obvious one—you must accurately align all the images to get a 3D reconstruction, which is very hard. Consider all the image artifacts and sample distortions that may arise during data acquisition, and it gets harder still.
Into the Future: A Perspective on vEM
If these are challenges, what’s next for vEM?
In a recent interview with us on The Microscopists, Lucy Collinson, Head of Electron Microscopy at the Francis Crick Institute, puts the future of volume electron microscopy like this:
The connectome map of the Caenorhabditis elegans worm’s brain exists thanks to vEM. [4] Now the effort of the connectomics community is directed toward providing the connectome of the fly brain. Once they have managed this, they will probably want to map the mouse connectome.
If the challenge of mapping the C. elegans mind is equivalent to one airline seat, then the fly brain is the equivalent of 4 Boeing 747 jumbo jets, and the mouse brain is the equivalent of the distance between Boston, MA, and Lisbon.
Lucy has also co-authored an excellent introduction to vEM [5] that goes into more detail than we do here.
The challenges in sample preparation, data acquisition time, image volume, and data analysis are enormous. Hopefully, the effort expended in achieving them will usher in the next generation of vEM technology, making it more accessible to every researcher.
References
- Motta A, Berning M, Boergens KM, et al. (2019) Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science 366:eaay3134
- Xu CS, Hayworth KJ, Lu Z, et al. (2017) Enhanced FIB-SEM systems for large-volume 3D imaging. Elife 6:e25916
- Glancy B (2023) MitoNet: A generalizable model for segmentation of individual mitochondria within electron microscopy datasets. Cell Syst 14(1):7–8
- Mulcahy B, Witvliet D, Holmyard D, et al. (2018) A Pipeline for Volume Electron Microscopy of the Caenorhabditis elegans Nervous System. Front Neural Circuits 12:94
- Collinson LM and Peddie CJ (2014) Exploring the third dimension: Volume electron microscopy comes of age. Micron 61:9–19







