You may be familiar with standard single fragment ligations: insert, vector, ligase, done! What if you have a complex cloning project with a massive region of DNA to clone? You can’t PCR the whole thing, and you can’t cut the entire thing out from somewhere else. What do you do?
Simple, you ligate multiple fragments into your vector all at the same time! Difficult, you say? Read on. We’ve got multiple fragment ligation covered! We’ll discuss the different methods and troubleshooting for when things go wrong.
Why would I Want to Perform a Multiple Fragment Ligation?
If you are trying to clone a large, say 7.5 kb coding sequence with difficult-to-PCR regions, you cannot amplify the whole thing. You likely have to amplify multiple fragments and stitch them together to create the final 7.5 kb insert encoding your favorite protein. Plus, you need to attach a specific strong promoter to said 7.5 kb insert. Sound familiar?
You might need to ligate multiple fragments into a single vector for many reasons, but I’ll guess your time is always in short supply. If you could perform a single ligation instead of three tedious sequential ligations and save time and the potential for scarring, all the better, right?
Sequential Ligation vs. One-step Ligation
Let’s cover the basics, starting with sequential ligations.
There are a few options for cloning methods, and this Bitesize Bio article on cloning methods is a good resource. One of the most common methods is using restriction enzyme ligation. You can also ligate the blunt ends if you cannot find unique or compatible restriction sites.
You can start by using PCR to amplify multiple pieces of your desired insert. Next, ligate the first amplicon into the vector, expand the construct, miniprep it, cut it, ligate the next amplicon, and so on. While it may be straightforward conceptually, this approach requires multiple ligations, transformations, minipreps, and intermediate QC steps to get to the final result. That is easily weeks of work.
Consider a multiple fragment ligation–one-step, days of work instead of weeks. What’s not to like? On paper, this is straightforward, but how do you ensure that you can match up compatible sticky ends? Or how do you calculate the molar ratios of inserts to the vector? What post-ligation QC steps will you need to ensure you have the complete construct, and how many clones will you need to screen?
Fortunately, several approaches have been developed to make multiple fragment ligations practical.
Gibson Assembly
The Gibson assembly method was invented by Daniel Gibson in 2009. [1] This method allows you to select overlapping regions between fragments, so there is no need to worry about compatible restriction sites or scarring.
Step 1: Generate the multiple fragments you are interested in cloning using PCR. Each fragment will contain an overlapping sequence with the next fragment in the series. The assembly will be directional. You can also prepare your vector at this stage, linearizing using restriction endonucleases as you would for any conventional cloning procedure.
Step 2: Post-PCR clean-up of amplicons to remove the PCR reagents and unincorporated nucleotides, and post-digest clean-up of your linearized vector.
Step 3: in a single-tube reaction, combine all purified linear fragments and the linearized vector. A 5’ DNA exonuclease erodes the 5’ end of all dsDNA to create sticky ends of varying lengths. The adjacent fragments will now anneal directionally, with a DNA polymerase to fill in any gaps and a ligase to seal the nicks. And now you have multiple fragments ligated in order into your vector! (Figure 1)
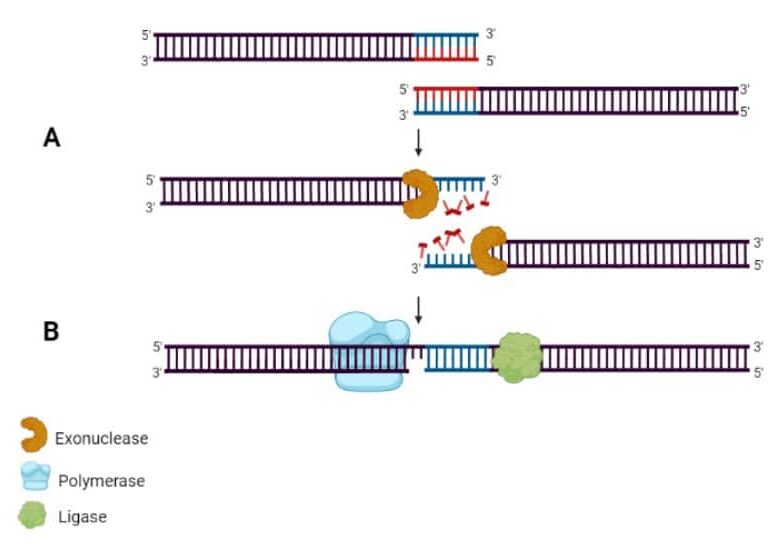
After creating the construct, you transform competent E. coli as you would for any ligation and screen the resulting transformants for the correct clone. You can check out this Bitesize Bio article on Gibson Assembly if you are interested in learning more.
Other Methods for Multi Fragment Ligations
The Gibson method is not the only multi-fragment assembly method. Below we will go into some alternatives giving you the pros and cons.
Using Conventional Ligation Reagents
You can try mixing multiple fragments and a linearized vector together in a single ligation reaction.
Your fragments can be generated individually by PCR, or you can purchase synthetic annealed oligonucleotides. Getting the different fragments is the easy part. To directionally ligate them, you must strategically incorporate restriction sites.
If the restriction sites are not present in the native sequence of each fragment, they can be engineered in, but your construct will now include some extraneous sequences. If you are trying to express what you are cloning, then careful consideration is needed to maintain the reading frame and what effect an extra amino acid or two will have on your favorite protein.
If convenient restriction sites are not present in your native sequence or you cannot engineer them in, you can try blunt-end cloning. This makes it easier to maintain your coding sequence in an expression construct and avoids incorporating extraneous sequences, but blunt-end cloning is non-directional. Mixing all fragments and your vector and relying on chance to get the correct fragment orientation will likely be next to impossible.
Selectively dephosphorylating 5’ ends can overcome the orientation problem with blunt end cloning and may be the best alternative if native restriction sites aren’t present.
Cloning Systems
Various other cloning approaches like the Gateway system or TOPO (TA) cloning are on the market to make single insert cloning easier. These systems use approaches to make ligations more efficient, and that certainly can be an asset.
However, the same issue you’d encounter directionally ligating multiple inserts still exists with these cloning systems. While the efficiency of ligation may exceed that of conventional cloning, you still must grapple with getting the multi-way ligation conditions just right. And that leads us to the next section!
Optimization
The conditions for a multiple fragment ligation must be optimized. Ligation conditions that work just fine for one insert may not favor the ligation of multiple inserts. Suboptimal ligation conditions could produce incomplete constructs lacking one or more of your intended fragments or uncontrolled concatenations of your fragments and the vector.
You can try trial and error, and maybe even a lot of trial and error! Or you may consider a Design of Experiments approach.
One interesting method developed by An et al. (2010) performs the ligation of multiple fragments first and then uses this as the input to a PCR. [2] There may be multiple ligation products, but you can amplify the correct one based on size by gel purifying the correctly sized fragment. In this case, it doesn’t matter if the fully ligated fragment is favored because PCR can amplify that needle in a haystack.
This is a unique way to isolate the multiple fragment ligation product and then just clone a single large insert into your vector. This method requires a bit of optimization, but if you get the conditions right, it is an efficient alternative to the Gibson method.
In general, ligation conditions must account for:
- Specific ligase selected and the amount used.
- Ligation time and temperature.
- Size of fragments—you may be constrained by where restriction sites occur within the native sequences of your target fragments.
- Molar ratios of fragments to the vector.
Tips and Tricks for Performing Multiple Fragment Ligations
Thanks in part to the growth of synthetic biology, you are not alone in facing the challenges of multiple fragment ligations! Commercial kits exist to make your life easier, and there is a lot of know-how on the web and in the literature. Here is a brief list to help you along.
Ligation Reagent | Functions |
PCR or synthetic oligonucleotides | Standard PCR reagents and oligo design techniques are used to amplify your desired fragments. |
Gel purification and PCR clean-up | PCR products and linearized vectors should be cleaned up before ligation. Gel purification or column-based clean-up procedures will yield purified amplicons ready for ligation. |
Restriction enzymes | Vectors are linearized using selected restriction endonucleases. |
Exonuclease, DNA polymerase, and ligase | These are required for the Gibson method, but these can be purchased in commercially available kits. |
Competent cells | Select competent cells with the desired efficiency for cloning. |
Miniprep and QC reagents | Any general reagents or kits for performing minipreps to isolate plasmid DNA from clones. |
Optimal conditions | Multiple fragment cloning can take some trial and error with respect to molar ratios of fragments comprising the insert and the vector, as well as times and temperatures for ligation and other manipulations. |
This kit by Takara takes advantage of a very efficient ligation enzyme that recognizes 15 bp overhangs engineered into your inserts using specifically designed PCR primers. | |
NEB provides a kit containing the enzyme mix for Gibson assembly as well as specially optimized E. coli for transformation. |
Multiple fragment ligations can be tricky, but there are a few methods and many tools and commercial reagents to make your cloning project a reality. Please leave a comment below if you have any tricks for multiple fragment ligations!
Need a helping hand calculating the amount of insert for your ligations? Download our free lab math cheat sheet for a handy formula and lots of helpful calculations.
References
- Gibson, DG et al. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345.
- An, Y et al. (2010) PCR-after-ligation method for cloning of multiple DNA inserts, Anal Biochem. 402: 203-205
- Revie, D et al. (1988). Kinetic analysis for optimization of DNA ligation reactions. Nucleic Acids Res. 16:10301–10321.