In biological research, we often try to mimic cellular events in the lab.
Phosphorylation is a common and fundamental cellular event without which life as we know it would not exist.
So, it’s probable that you will need to phosphorylate a protein during your research.
But what is phosphorylation, what amino acids get phosphorylated, and how can we mimic it in the lab?
In this article, we’ll answer all those questions. Even better, we’ll discuss phosphomimetics that work in vitro and in vivo.
Why Phosphorylate a Protein?
Why would you ever need to phosphorylate a protein or mimic phosphorylation in a protein?
Good question. Examples that sprang to my mind include:
- To manipulate biological systems in vivo. (e.g., lux reporter assays.)
- To manipulate biological systems in vitro. (e.g., enzyme kinetics.)
- To check if a purified protein sample is folded and active.
- To prepare positive and negative controls for assays.
- To prepare physiologically-relevant samples for structural studies.
- To investigate the functional importance of phosphorylation at a given site.
For example, you could use it to show sample oligomerization upon phosphorylation.
Or perhaps demonstrate increased or decreased binding affinity to a co-factor upon phosphorylation.
Alternatively, you might want to explore or prove that ligands bind to your target only when it is in a phosphorylated or non-phosphorylated state.
You might want to rule out phosphorylation as a variable to isolate the effect of other variables. Imagine, for example, that you want to explore the influence of disulfide bridge formation on your sample’s activity.
You could exploit phospho-mimetics to lock your sample into a phosphorylated or non-phosphorylated state and then prepare point mutants to abolish said disulfide bridge.
Maybe you can think of a few more?
What Is Phosphorylation?
Phosphorylation is a post-translational modification where an orthophosphate anion is covalently attached to an amino acid side chain. [1]
It occurs in bacteria, archaea, and eukarya.
Examples of cellular processes that phosphorylation assists in regulating include:
- Cell-to-cell signaling.
- Signal transduction.
- Gene regulation.
- Apoptosis.
The addition of a phosphate anion to the surface of a protein modifies its function so that it can assist in carrying out a given process.
The main ways that phosphorylation modifies protein function are:
- Inducing structural rearrangement.
- Providing chemistry (a high-energy P-O bond) for downstream processes.
- Lowering its isoelectric point.
Regarding point 1, structural rearrangement may lead to oligomerization. This is frequently observed for response regulator proteins. In such cases, the response regulator usually dimerizes upon phosphorylation to bind to a pair of linear DNA repeats.
Regarding point 2, the phosphate anions are covalently linked to amino acid side chains through a [phosphate]-P-O-[residue] bond. These are comparatively weak and susceptible to both enzymatic cleavage and hydrolysis.
According to Gibbs free energy, releasing energy from a chemical bond makes a process more thermodynamically favorable.
And because the P-O bond is comparatively weak, spontaneous dephosphorylation can occur via hydrolysis.
Regarding point 3, phosphate is the conjugate base of phosphoric acid and is negatively charged at physiological pH. So, when added to a protein, the overall proportion of the negative charge on the protein is increased. That’s to say, the protein is effectively made more acidic.
The types of functional changes that are induced by phosphorylation include:
- Promoting binding to a partner.
- Altering intrinsic stability to induce proteasomal degradation.
- Direct activation or inactivation of enzymes.
- Changing localization.
Which Amino Acids Get Phosphorylated?
A non-exhaustive list of amino acids that are capable of being phosphorylated include:
- Serine.
- Threonine.
- Tyrosine.
- Arginine.
- Aspartic acid.
- Histidine.
- Cysteine.
Phosphorylated serine, threonine, and tyrosine are more common than the others on the list. In these instances, a phosphate replaces the side chain hydroxyl group (OH).
The diversity of phosphorylatable amino acids is surprising and encompasses positively charged, negatively charged, and hydrophobic residues.
3 Ways to Mimic Phosphorylation
You can mimic phosphorylation genetically and chemically. So, it doesn’t matter whether you want to conduct your experiments in vivo or in vitro—there’s a method for you!
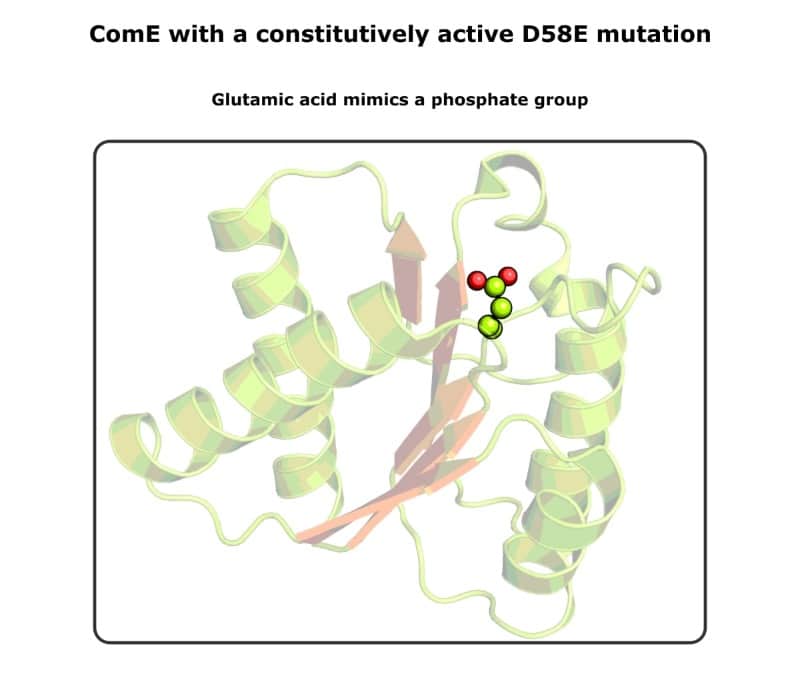
1. Prepare Aspartic or Glutamic Acid Point Mutants
Single-site point mutants to aspartic or glutamic acid can mimic phosphorylation. [2]
This mimicking is reasonably well documented in the scientific literature if not totally understood. Presumably, the negative charge on the side chain is a sufficiently good mimetic of the phosphate anion to produce the desired effect most of the time.
So, use inverse PCR to mutate the residue that gets phosphorylated to aspartic or glutamic acid. With a bit of luck, you should have a constitutively active or inactive protein for your assays or structural studies!
Remember, constitutive means having basal activity without a ligand.
Agilent has a helpful tool to help you design primers if you’re new to site-directed mutagenesis.
Because you are altering the DNA sequence rather than biochemically modifying a protein sample, this method works great in vivo.
Nothing is stopping you from ligating the mutated gene onto an overexpression vector and purifying the constitutively active protein. This approach is used routinely for structural studies because the change is permanent. Check out Figure 1 for an example.
You can also use unnatural amino acids to mimic phosphorylation if you wish. Check out this paper by Gonzales et al. for a handy table of unnatural amino acids for mimicking phosphorylation. [3]
Prepare a Non-Phosphorylatable Control
Science is nothing without a good set of controls.
You can use the site-directed mutagenesis approach to prepare non-phosphorylatable controls and samples. Precisely what a non-phosphorylatable protein achieves depends on what property phosphorylation imparts in the system you study.
Simply mutate the phosphate-receiving residue to either glutamine or alanine. If the phosphate-receiving residue is a tyrosine, mutate it to a phenylalanine residue.
Glutamine or phenylalanine may represent more conservative mutations if the wild-type residue is glutamic acid or tyrosine, respectively. This is because they are structurally similar to the suggested mutants.
And alanine is absent of any charge. It’s your choice. Check out Table 1 below for a summary of beneficial mutations.
Table 1: Mutations to mimic or prevent phosphorylation.
Phospho-mimetic mutations | Non-phosphorylatable mutations |
Aspartic acid | Alanine |
Glutamic acid | Glutamine |
Phenylalanine |
Things That Can Go Wrong
Anticipate:
- The PCR might fail.
- Your mutants might be toxic to your cells.
- You might not be able to transform your mutants.
- Mutants may not mimic phosphorylated or unphosphorylated residues.
These points are all related. The intricacies of the system you work on will dictate what specific problems you may encounter.
For example, if you overexpress a constitutively active response regulator to high levels, it could indiscriminately switch on non-cognate genes in the host cells. This could lead to points 2 and 3.
I’ve experienced one occasion where a native host inserted an RNase gene into a particularly toxic plasmid. Only when I mini-prepped it, discovered it was mysteriously a few kilobases larger than expected, and got the extra bit sequenced did I find out.
My target gene was snipped to ribbons as it was expressed. Ouch!
2. Use Chemical Phospho-Donors
You can buy phospho-donors off the shelf and add them directly to a protein sample to phosphorylate it. [4] Common phosphorylating agents include:
- Acetyl phosphate.
- Phosphoramidate salts.
- Carbamoyl phosphate.
- Sodium pyrophosphate.
Note that these are high-energy compounds. This has two consequences:
- They have short half-lives in aqueous solutions.
- The phospho-derivates they form are impermanent.
Regarding point 1, check with the manufacturer for stability information. If you can’t find any relevant information, assume it has a half-life of approximately one hour.
Regarding point 2, the stability of the phosphate derivative of your target depends on the target itself. If you have a robust assay, you can determine its stability by conducting a time course and measuring how long it takes for whatever change is imparted by phosphorylation to wear off.
Because of both these reasons, these donors are seldom used for structural studies.
Things That Can Go Wrong
Anticipate:
- Your donor/derivate may hydrolyze before you assay your protein.
- You’ll need to determine what concentration of donor to add.
Regarding point 1, work at ice-cold temperatures. This should prolong the lifetime of unstable species.
Regarding point 2, an activity assay should enable you to determine an appropriate donor concentration. Otherwise, start with several molar equivalents.
3. Prepare Your Own Phosphate Mimetic
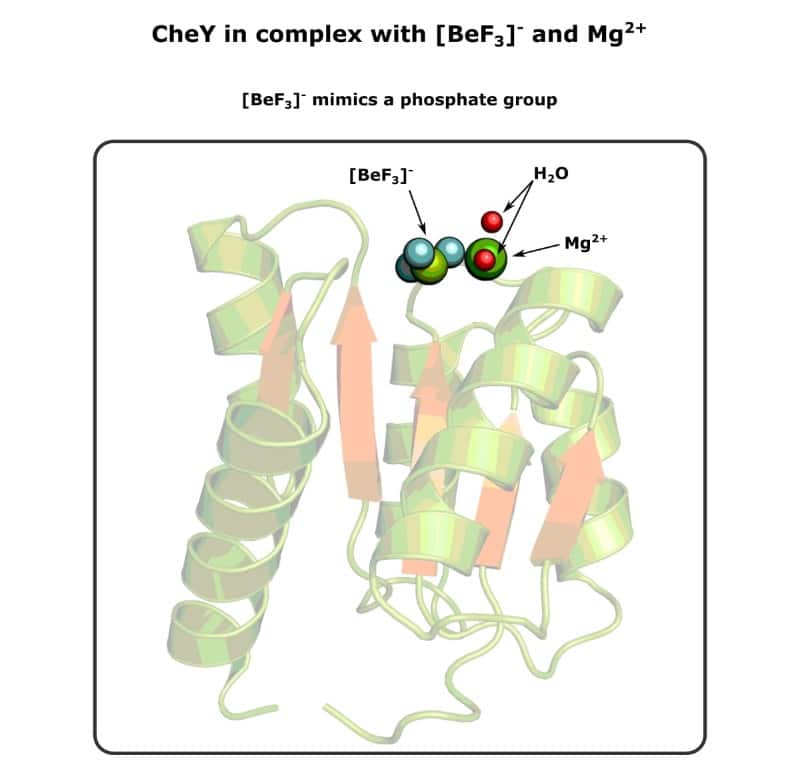
Beryllium trifluoride (BeF3–, also called beryllofluoride) is a phosphate mimetic that you can prepare yourself. [6–8]
Do so by mixing:
- 15 mM sodium fluoride (NaF).
- 3 mM beryllium chloride (BeCl2).
- 3 mM magnesium chloride (MgCl2).
Wait half an hour, and hey presto—your own mimetic. If you need a stock with a higher concentration, elevate these concentrations but keep their proportions.
It’s not strictly a phospho-donor because there’s no phosphate group (or even a phosphorus atom).
Oh, and you likely get a mixture of beryllium adduct products. So, you’ll often see it denoted as BeFx.
BeF3– structurally and chemically resembles the tetrahedral orthophosphate anion (although it is trigonal pyramidal like ammonia).
So, it does a great job of mimicking phosphate. Once you’ve prepared it, treat it just like the phospho-donors listed already.
It has a few other excellent properties that are worth noting:
- It’s stable in aqueous solutions.
- It binds ionically to proteins.
- It binds irreversibly to proteins.
So, there’s no need to worry about hydrolysis.
Usually, it binds to proteins in tandem with a divalent metal such as Mg2+.
And because it’s a potent nucleophile, it’s comparatively harsh and may precipitate your sample.
Things That Can Go Wrong
Anticipate:
- Beryllium trifluoride may precipitate your protein.
- Negative results could be ambiguous.
Regarding point 1, this is because beryllium trifluoride is a harsh chemical. If the protein you are studying binds to co-factors or DNA, etc., try adding these before adding in beryllium trifluoride because they might protect your sample protein.
Even if this means reversing the order in which events occur in vivo.
For example, I once worked on a response regulator exhibiting increased affinity for promotor DNA upon phosphorylation—adding beryllium trifluoride before DNA precipitated the protein, however.
I overcame this by adding excess DNA first and then adding beryllium trifluoride. The mysteries, eh!
Regarding point 2, this is because if you prepare your own beryllium trifluoride. And if your activity assay gives you a null result, you cannot conclusively prove that you successfully prepared beryllium trifluoride and rule it out as a reason.
When we mix beryllium and fluoride salts, we trust that beryllium trifluoride spontaneously forms.
And it does work because people prepare it routinely. At the time of writing, there are approximately 500 entries in the PDB with beryllium trifluoride in the deposition title, and I’ve drawn an example below in Figure 2.
But ultimately, the burden of proof lies with you.
Phosphomimetics Summarized
We’ve covered quite a bit of ground in this article. So, see Table 2 for a quick summary of the three methods discussed.
Table 2: Pros and cons of methods for mimicking phosphorylation.
Method | Pros | Cons |
Mutagenesis | Permanent | Mutations may be toxic to the host |
Chemical phospho-donors | Closely resembles physiological phosphorylation | Impermanent because Phospho-donors are unstable |
Beryllium trifluoride | Permanent | May precipitate your protein in vitro |
Ways to Mimic Phosphorylation Summarized
So, there we have it. Mutagenesis, chemical phospho-donors, and beryllium trifluoride. Three easy methods to phosphorylate proteins. These should cover you, whether you work on cells, solutions, or both.
Plus, a heap of other information to help you design your experiments! (And good luck with those.)
And please note that the ways to mimic phosphorylation are by no means exhaustive. Reference 2 makes for excellent further reading on the topic.
Have I missed your favorite phospho-donor off the list? Got any pearls of wisdom about these fiddly but beneficial procedures? If so, get in touch or drop a comment below.
References
- Pawson T and Scott J (2005) Protein phosphorylation in signaling – 50 years and counting. Trends Biochem Sci 30:286–90
- Chen Z and Cole PA (2015) Synthetic approaches to protein phosphorylation. Curr Opin Chem Biol 28:115–22
- Pérez-Mejías G et al. (2020) Exploring protein phosphorylation by combining computational approaches and biochemical methods. Comput Struct Biotechnol J 18:1852–63
- Boudes M et al. Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae. Nucleic Acids Res 42:5302–13
- Lukat G, McCleary W, Stock A, and Stock J (1992) Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. PNAS 89:718–22
- Klose K, Weiss D, and Kustu S (1993) Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J Mol Biol 232:67–78
- Mesmer R and Baes C (1969) Fluoride complexes of beryllium(II) in aqueous media. Inorg Chem 8:618–26
- Gulik A et al. (2000) X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs. J Biol Chem 275:398–408
- Guhaniyogi J, Robinson V, and Stock A. (2006) Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J Microbiol 59:624–45