Site-directed mutagenesis (SDM) is a technique used to mutate one or more bases within a plasmid. This approach can change amino acid composition, destroy transcription factor binding sites, or create fusion proteins—to name a few examples.
Although SDM is most widely used to probe the structure and biological activity of nucleic acids and proteins, it is also useful for introducing or removing restriction enzyme recognition sites to aid cloning.
Furthermore, SDM can be a useful codon optimization tool to replace rare codons in expression vectors to improve heterologous protein expression efficiency. This is sometimes necessary because of codon bias, a phenomena whereby organisms have a preference for certain codons over others, depending on the availability of tRNAs in the cell. You can read more about codon usage and codon bias here.
When employed to change the coding sequence, SDM creates specific mutant constructs that lack critical residues or protein domains involved in post-translational modifications or regulation of protein stability. Removing critical residues or regions in your plasmids, such as phosphorylation sites or critical domains, provides one way to demonstrate the importance of certain protein features. For example, altering phosphorylation sites, such as serine to un-reactive alanine or to phosphomimetics such as aspartic acid, yields reliable evidence for site-specific protein phosphorylation. It allows the study of in vitro implications of such post-translational modifications.
Traditional Approaches to Site-Directed Mutagenesis
Inverse PCR
For deletion or insertions of >50 bp, inverse PCR is the most popular approach. Inverse PCR uses back-to-back primers to amplify the whole plasmid, followed by ligation of the linear product forming circular DNA. This technique is also suitable for larger insertions or deletions, e.g. removing a regulatory domain from a protein.
For deletions, the selected area can be removed by designing primers that anneal at either side of the targeted deletion zone. Amplification proceeds outwards from this area, thus excluding this region from the PCR product. Once the linear product is ligated, your new construct will be lacking this deletion domain. This process is outlined schematically in Figure 1 below.
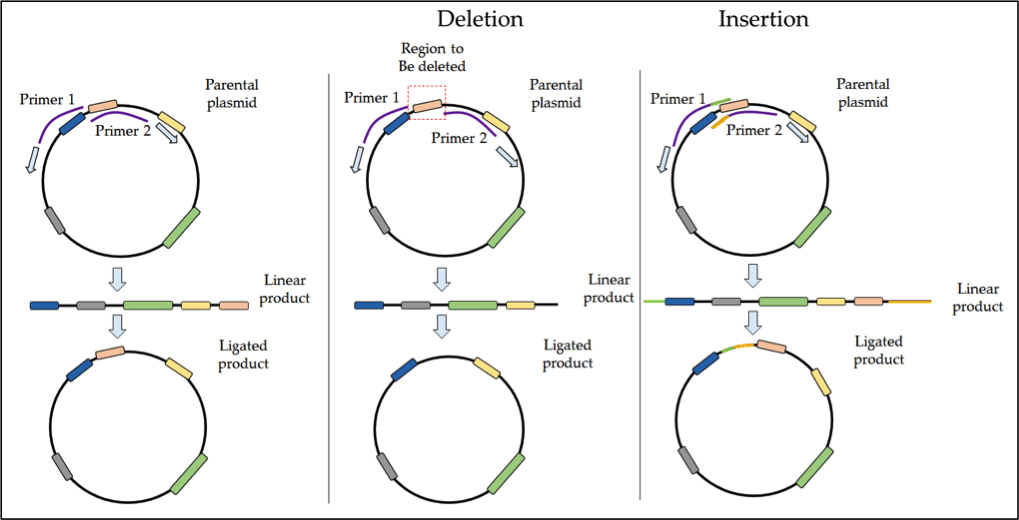
Figure 1. Protocol for inverse PCR in SDM.
Insertions can be achieved by using overlapping primers with flanking sequences that contain the appropriate extra bases. For more information on inverse PCR check out these resources:
- Ochman et al. (1998) Genetic Applications of an Inverse Polymerase Chain Reaction. Genetics. 120:621-623.
- Clifford N. Dominy and David W. Andrews. Site-Directed Mutagenesis by Inverse PCR.
SDM Using Modified Primers
This technique uses modified primers to incorporate small base pair changes into a plasmid, and is the method of choice for site-specific mutations. To start with, you’ll need to design some primers. But before you start:
- Print out a copy of an amino acid codon table
- Have your plasmid or protein sequence at the ready.
- Make sure you know where the start codon is and in what reading frame your sequence is read.
Primer Design
Aim for SDM primers of approximately 30 bp in length with your mutated site as close to the center as possible. While it is acceptable to make primers a little longer or shorter as required, there should be a minimum of 12 bp either side of your mutated site.
If you need help with primer design, https://molbiol-tools.ca/PCR.htm lists some really useful resources to set you on your way.
Pick the Right Reagents
Ordinary Taq polymerase just won’t cut it when it comes to SDM – you need to use a proofreading enzyme. There are a variety of commercially available polymerase kits that are up to the job, incorporating a range of features including high fidelity (proofreading capability) and hot start activation. One possibility is Takara’s In-Fusion HD Cloning Plus – an all in 1 solution that includes a high-fidelity polymerase, a PCR purification kit, cloning enzyme, and competent cells for site-directed mutagenesis.
Bear in mind that like most lab reagents, many polymerases come with their own pros and cons – generally labs will have historic reasons for picking one over the other and rarely stray from this. While sticking with what works is sensible, there is no harm in reading the literature on other available resources to make sure you have the best reagents for the job!
PCR Reaction
The PCR conditions used will vary depending on your choice of kit and polymerase, as well as the primers you have designed and the size of the product. However, the example shown below is a good starting point.
| Step | Temp (°C) | Time (s) | Cycles |
|---|---|---|---|
| Denature | 94 | 15 | 18 |
| Anneal | 60 | 30 | 18 |
| Extension | 72 | 20/kb plasmid | 18 |
Table 1: Example of SDM thermal cycling program
Keeping the number of cycles low will prevent unwanted mutations from occurring. Eighteen cycles should yield a reasonable amount of mutated product without incorporating unwanted mutations. Note that the number of cycles can be altered during troubleshooting if required.
Digest It!
Now that you have your PCR product ready, don’t forget the critical step – digestion! Here, you digest the parental template DNA to ensure that you only have the mutated plasmid for bacterial transformation. Standard protocols call for a 1 hour incubation at 37°C with endonuclease Dpn1 to digest all dam-methylated and hemi-methylated parental DNA, leaving you with your desired mutated plasmid.
Transform and Sequence
Transform your plasmid into competent cells just as you would with any other expression plasmid, and isolate single colonies for plasmid isolation.
Now you should have a small volume of your desired plasmid. Before you start experimenting, send a sample for sequencing to confirm the presence of your desired modification(s). It is equally as important to ensure the absence of any secondary undesired mutations.
If the sequencing returns with a positive result – congratulations, you are an SDM pro! But if the sequencing returns with a negative result – don’t fret! You can always try it again – our expert tips for troubleshooting SDM should get you back on track!
Nowadays, decreasing costs of oligonucleotide synthesis and advances in synthetic biology means synthetic approaches are gaining traction over site-directed mutagenesis. Furthermore, the emergence of CRISPR/Cas9 technology has also simplified gene editing such that mutagenesis can now be performed in vitro and in vivo in a few simple steps. Nevertheless, SDM continues to be a mainstay in the molecular biology toolbox, and is not going anywhere anytime soon.
Originally published in 2016. Updated and republished in 2018.







