You’ve no doubt heard of flow cytometry, the go-to method to analyze heterogeneous cell populations at a single-cell level. But, hey! Do you want to know about a cool way to detect and tell the difference between virus particles? Have you ever heard of flow virometry?
If you haven’t, you’ve come to the right place! We’re going to take you through what this technique is, its applications and relevance to your biological research, and how you can start using it in your experiments.
Current Viral Detection Methods: Pitfalls and Problems
Commonly used assays to detect virus particles include plaque assays and enzyme-linked immunosorbent assays (ELISA). Western blots are often used when we need to analyze the expression of the viral proteins, but they can be cumbersome.
While plaque assays and ELISA are gold-standard and work great, they’re not as flexible when it comes to multiplexing an experiment. For example, a captured antibody (only one) is coated on one ELISA plate at a time. And as for Western blotting, multiple proteins can be detected, but this involves stripping the original antibody and re-probing for a different protein.
It’s clearly time for a new method to rapidly quantify and analyze viral particles in their native form and in real time.
Flow Virometry: What Can It Do for You?
Basically, flow virometry is a specialized way to characterize and analyze individual viral particles using a flow cytometer.
A flow cytometer can fetch your samples suspended in fluids and count each individual particle (cells and viruses included). But it can do so many cool things beyond counting.
For example, it can tell the difference in size and level of the viral protein expressed on the surface of the viral particle.
An average virus particle size is super tiny! It ranges from 17 nm to 350 nm, [1] and that’s a thousand times smaller than a dust particle.
Therefore, you need to fine-tune your flow cytometer and prepare the samples carefully to get the best results. Check out our related article on how a flow cytometer works for a more in-depth explanation, or else read on (Figure 1).
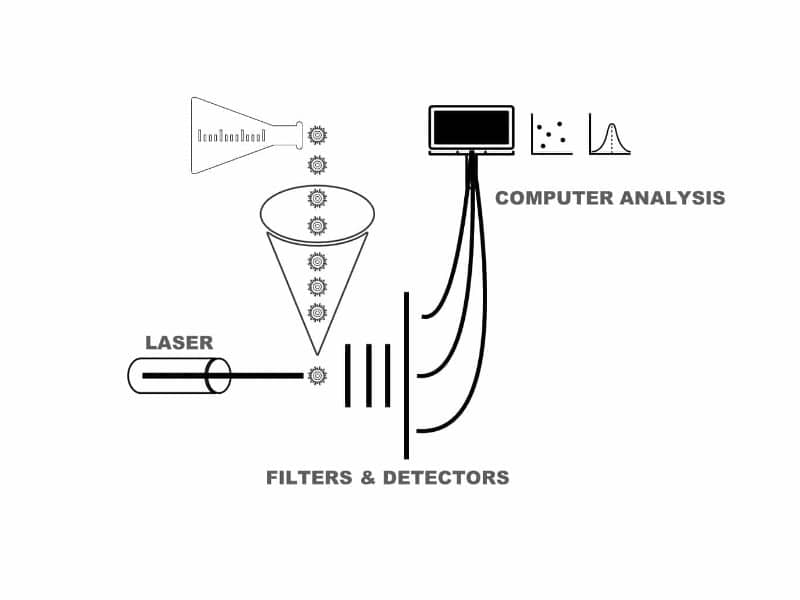
How a Flow Cytometer Works
A. The Fluidic System
Your sample is taken up by the fluidic system through a series of tubes until it reaches a narrow passageway where each individual particle can be focused.
B. The Optical and Detector System
This includes different lasers, optical filters, and detectors. The optical filters are selective in the wavelength of the fluorescence signal being passed through. The fluorescence signals are then converted to electrical signals through detectors.
C. The Data Acquisition System
This final part stores all the digital signals of each particle for further analysis.
How Can You Get Started?
Know Your Cytometer
Not all cytometers are built the same. Some mini cytometers are geared towards counting red blood cells, and some are equipped with cell-sorting ability. This expensive equipment requires great precision in operation. So please consult your flow facility manager on how to set up the machine.
The first thing you need to figure out is what type of flow cytometer you’ll be using. Most flow cytometers are calibrated to detect cells (ranging from 10 mM to100 mM in diameter). I’m just going to briefly talk about how to tune your typical flow cytometer to detect viral particles.
First Modification: Forward Scatter Setting (FSC)
When a light beam strikes a virus particle at a 90-degree angle, the light that passes through the particle is refracted, and the FSC angle can be recorded. A typical FSC is usually set at an angle of 0.5–15 degrees.
For microscopic particles that scatter light at a wider scattering angle, next-generation cytometers need to set the detection angle at 15–70 degrees (a reduced wide-angle detection).
Second Modification: Increase the Laser and Detector Power
The power of the light source (lasers) also needs to be cranked up a notch or two. This is because the average size of a virus particle is smaller than the wavelength of the light source, and, therefore, it refracts the light source poorly. [1]
In addition, flow cytometers should be equipped with high-performance photomultiplier tubes (PMT) rather than the traditional photodiode detectors, [1] which significantly increases their detection sensitivity.
The methodologies described by Tang et al. can provide a great starting point for learning how to optimize your laser voltage. [2]
Third Modification: Filtration! Filtration! Filtration!
Filtration of the buffer is essential to minimize the background noise in your samples. To minimize the background noise generated by impurities such as crystals, or microscopic bubbles in the buffer system, the buffer system used for flow virometry should be pre-filtered with 0.1-mM filters.
Having said that, all the virus sample stocks should be pre-filtered with 0.45-mM filters to prevent the inclusion of cellular debris and protein aggregations.
Final Considerations
Decreasing the flow rate has been shown to increase the signal-to-noise ratio for detection in general.
Labeling of the Virus Particles
To really identify the virus that you want to study, you might want to consider antibodies that will recognize your virus of interest to tell the virus particle apart from any background noise (i.e., cell, debris, exosomes).
Fluorescence-labeling of the virus population with different kinds of probes is highly recommended. Below I’ll cover some common methods for fluorescence labeling your virus population.
Virus-Specific Monoclonal Antibodies for Surface Labeling
A straightforward method is to get a monoclonal antibody that specifically recognizes the virus surface protein you are interested in studying. Just imagine these antibodies as tags for a specific viral surface antigen.
The viral antigen should be abundant, and the monoclonal antibody can either be directly labeled or be labeled with a fluorescence molecule via a secondary antibody.
Virus-Specific Antibodies Pre-Bound to Nano-beads can Improve Light Scattering
One approach is first to use nanoparticle-coupled antibodies to capture the virus, followed by virus-specific fluorescence staining. [3]
One of the great advantages of this method is that you can enrich the virus populations by using magnetic nano-beads coupled with virus-specific antibodies. However, one caveat when using nano-beads is that multiple viruses might be attached to one bead, making single-particle analysis impossible.
Labeling of Viral Nucleic Acids
This method might be better for DNA viruses because they have larger genomes. A large sample stock of DNA viruses can be fixed and stained with a common DNA dye such as SYBR Green. [4] An RNA virus might prove more challenging for nucleic acid staining because of the low copy numbers and small genome size. [4]
Lipophilic Dyes for Enveloped Viruses
For enveloped viruses, you can try to stain their lipid bilayer membrane with lipophilic dyes. Experiments have successfully used DiD, DiO, and Dil dyes for HIV, vaccinia, and dengue. [4] However, contamination from exosomes and micro-vesicles is always a concern.
GFP-Viruses
If you have genetically engineered viruses that express fluorescent molecules such as green fluorescence protein (GFP), it will definitely make your life much easier for tracking. In addition, GFP-viruses of different sizes could come in handy when you are trying to calibrate your machine for flow virometry if fluorescent beads are not available commercially.
One major caveat of labeling viruses this way is that the manipulation with either antibodies or dyes could affect the viruses’ ability to infect or replicate properly, so consider this move carefully if you’re planning downstream experiments.
Controls for the Experiments
Control experiments are essential for you to conclude your analysis. Here are some recommendations for analyzing your virus of interest using flow virometry.
The flow cytometer needs to be recalibrated to analyze virus particles. To ensure that this has been done correctly, you can use commercially available synthetic micro-beads of various sizes to ensure that your machine can differentiate the microscopic molecules.
Virus titration should be performed to ensure the optimal range for FSC and fluorescence detection.
Traditional virus bioassays such as plaque assay, ELISA, or RT-PCR can be run in parallel to compare with the results from flow virometry.
Applications
So, if we can successfully track our viruses using flow virometry, what are the applications of this technique? Let’s take a closer look at some applications.
Detection of Single Viral Particles to Study Discrete Viral Particles and Viral Surfaces
Do you want to detect all the exotic microbes that live in your neighboring lake? Not only is flow virometry great for ecological studies, but it is also useful for public health screening of harmful pathogens. For example, we might need to prepare a fast and easy way to screen for any potential pathogens that could lead to a disease outbreak.
Flow virometry can open up new avenues for rapid virus detection in clinical settings as well. The high-throughput nature of flow virometry means that we can screen for potential viruses in different patient samples quickly.
There has already been a proposal to use flow virometry for rapid screening of SARS-CoV-2. [5] This rapid screening method could work together with the widely used, highly sensitive RT-PCR method to achieve great results.
Flow virometry can enable you to rapidly detect and characterize the same virus across batches in laboratory settings. For example, you can investigate how different levels of viral surface antigen expressions could relate to a virus’s level of infectivity. [6]
Virus Sorting and Discerning Between Viral Populations or Subtypes
With a proper flow cytometer, you can also sort different virus populations within a single batch. We all know that even within a single batch, each virus particle is different in the surface protein complement and genetic contents.
For example, you can investigate how these variations in virus surface protein number and genome structure can play important roles in infectivity and life cycle by sorting them into different populations.
In addition, to increase the purity of the virus stock, pre-sorting with virus-specific magnetic beads can be used. An HIV study utilized this method to purify and differentiate the HIV-particle (CD45– from CD45+) exosome populations using a regular flow cytometer. [7]
Virus Quantitation
Quantitation of any virus is important in both laboratory and clinical settings. Traditionally, cell-based assays such as plaque assays are used to estimate the number of infectious viral particles per ml. Depending on the type of virus, different incubation times are required for plaques to form on a cell monolayer.
Other methods such as ELISA or RT-PCR can easily take up to a day to perform. Therefore, we need a high-throughput assay for the detection and quantitation of viral particles. Well, step in flow virometry! Scientists have successfully quantitated marine and respiratory viruses using this technique. [8,9]
However, one caveat of this application is that flow virometry might not differentiate between an infective and a non-infective virus.
Vaccine Quality Assurance
For those of you who are screening for viral inhibitory compounds, do you ever wonder why the viral inhibitory compound activity varies depending on a particular batch of viruses you are using? Flow virometry may be able to help you in addressing the quality as well as quantity difference between virus batches.
For pharmaceutical companies, ensuring that the quality of different batches of vaccines remains consistent is paramount. Flow virometry is a potential high-throughput solution for monitoring the quality of your virus stocks. [10]
What’s Next?
Flow virometry is an exciting tool for the rapid detection and characterization of viruses. In clinical settings, it has excellent potential for providing rapid and high-throughput detection of virus particles. In laboratories, it has utility in the quantitation and characterization of viruses for biological research. Its high-throughput capability is an excellent screening and quality-control tool for different virus stocks. In addition, different virus populations can also be sorted by their unique characteristics. It is truly an exciting time to study viruses.
Considerations and Caveats of Flow Virometry
While flow virometry as a technique has great potential, some caveats still need to be addressed.
First, the fact that we look at single virus particles typically at a nanometer scale requires very delicate tuning of the flow cytometer and stringent background controls. For example, the fluidic system needs to be flushed and filtered with 0.1-mM filters to ensure the lowest level of microparticle contamination. [1]
Aside from that, all the virus samples need to be filtered with 0.45-mM filters to remove cellular debris and other contaminants.
Second, in addition to viral particles, micro-vesicles often exist in a virus stock. How do you differentiate between them since they are roughly similar in size? Here, fluorescent staining of specific markers on vesicles becomes a key issue.
Finally, how can we design next-generation flow cytometers equipped with better fluidics, more powerful lasers, and improved detectors to lead us into the era of nanoparticle flow cytometry?
Scientists are already considering the use of mass cytometers to study viruses. [1] A new semiconductor-based flow cytometer has also been developed with an ultra-sensitive detector. [11]
With the continued emergence of infectious diseases, there is an urgent need to develop a high-throughput method to detect and characterize every infectious agent. Flow virometry is well-suited for our future research needs. I can’t wait to find out what future discoveries about viruses are made using flow virometry!
Do you use flow virometry in your research? Get in touch with us in the comments if so!
References
- Lippe R (2008) Flow virometry: a powerful tool to functionally characterize viruses. J Virol 92(3): e01765-17.
- Tang V (2017) Single particle discrimination of retroviruses from extracellular vesicles by nanoscale flow cytometry. Sci Rep 7:17769.
- Yan X et al. (2005) Multiplexed flow cytometric immunoassay for influenza virus detection and differentiation. Anal Chem 77(23): 7673–8.
- Reyes Zamora JL et al. (2018) Flow cytometry as a tool to study viruses. Methods 134–135: 87–97.
- Soni N et al. (2020) A flow virometry process proposed for detection of SARS-CoV-2 and large scale screening of COVID-19 cases. Future Virology 15(8):525–32.
- Renner TM et al. (2020) Intact viral particle counts measured by flow virometry provide insight into the infectivity and genome packaging efficiency of Moloney Murine Leukemia Virus. J Virol 94(2): e01600-19.
- Bonar MM et al. (2017) High sensitivity detection and sorting of infectious human immunodeficiency virus (HIV-1) particles by flow virometry. Virology 505:80–90.
- Ricci G et al. (2021) Flow virometry for process monitoring of live virus vaccines-lessons learned from ERVEBO. Sci Rep 11:7432.
- Marie D et al. (1999) Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol 65:45–52.
- Ferris MM et al. (2002) Rapid enumeration of respiratory viruses. Anal Chem 74: 1849
- Brittain IV et al. (2019) A novel semiconductor-based flow cytometer with enhanced light-scatter sensitivity for the analysis of biological nanoparticles. Sci Rep 9: 16039.





