In 2008, Osamu Shimomura, Martin Chalfie, and Roger Y. Tsien won the Nobel Prize in Chemistry for “the discovery and development of the green fluorescent protein, GFP.” Nobody was surprised, as using fluorescent proteins revolutionized protein studies. Fluorescent proteins enabled the transition from the era of molecular biology—recombinant DNAs—to the era of cellular biology: studying biomolecules in vivo. But have you ever wondered how to visualize that notoriously difficult molecule, RNA? In this article, we’ll show you how, with light-up RNA aptamers.
The Problem of Visualizing RNA
It’s tricky to use the same approach of fusing reporter and target molecules of RNA because they’re much more unstable than proteins.
Of course, there are ways to find out about RNA dynamics. The number of specific RNA molecules in a sample can be studied with RT-PCR, for example. The cellular RNAs can be seen in tissue using a modified fluorescent in-situ hybridization (FISH) for RNA—ISH—by hybridizing fluorescent oligonucleotide probes to the target RNAs. But ISH requires tissue fixing with associated artifacts. And it doesn’t allow for the study of the dynamic processes of RNA metabolism, as fluorescent proteins can do for proteins.
Introducing Fluorescent RNA Aptamers
Luckily, there’s another method to study RNA dynamics—fluorescent RNA aptamers. Aptamers are oligonucleotides that fold into specific tertiary structures that can bind specific targets.
Add a fluorescent tag to the end of an aptamer, choose mRNA as a binding target, and you have something like FISH, but working in live cells. Aptamers can also work as stand-alone sensors of metabolites or tag ends of RNA molecules.
What Is a LURA?
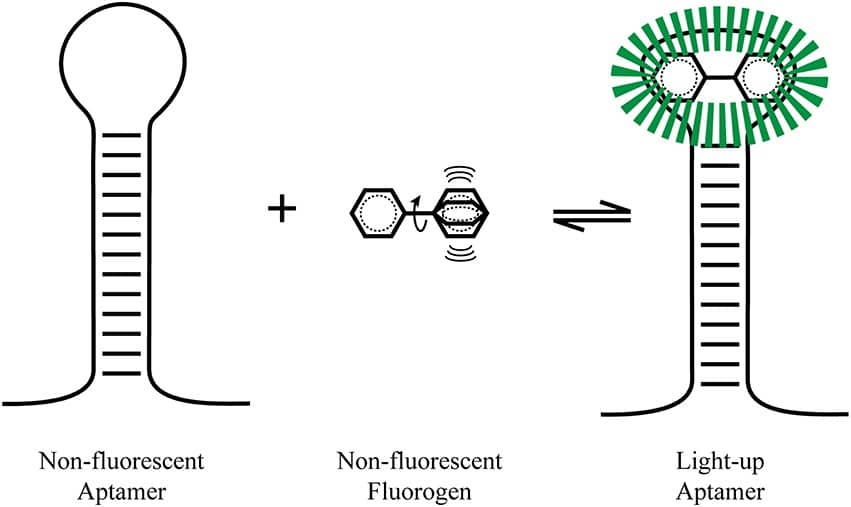
Light-Up RNA Aptamers (LURAs) are endogenous RNAs that fluoresce after binding a non-fluorescent small molecule. Binding to the aptamer channels the molecule’s energy that dissipates in the free state into fluorescence (Figure 1). [1]
Examples of LURAs include Hoechst dye derivatives that bind to a 29-nucleotide-long RNA blue fluorescent aptamer. [2]
How are RNA Aptamers Attached?
Aptamers can be attached to the ends of almost any RNA molecule and don’t interfere with their function. Aptamers start to function immediately after transcription, which precludes the steps that protein tags go through before becoming functional, namely translation and folding.
Selecting New RNA Aptamers with SELEX
New aptamers are obtained from “systematic evolution of ligands by exponential enrichment” (SELEX).
In brief, SELEX is an extensive oligonucleotide library consisting of randomly generated sequences flanked by constant 5′ and 3′ ends. These ends work as primers. The library sequences are mixed with the target molecule, and the molecules that don’t bind the target are removed by affinity chromatography or target capture on paramagnetic beads.
Finally, the bound oligonucleotides are eluted and amplified by PCR for subsequent rounds of selection.
What Can I Study with Light-up RNA Aptamers?
You can use LURAs to ask many research questions, on topics ranging from metabolism to translation. You just need to be creative and read the literature. For example:
- LURAs can function as riboswitches. Riboswitches are parts of RNA that fold in a stable structure and change the fluorescence after binding a metabolite, such as the important enzyme cofactor S-adenosyl-methionine (SAM). [3]
- LURAs can be used to study RNA synthesis and trafficking. [4,5]
- LURAs can be used for intracellular viral DNA dynamics. [6]
Currently Available Aptamers
One of the first aptamers that was developed binds a green fluorophore: 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), and is imaginatively named “Spinach.” (Confusingly, the applications that are listed above have nothing to do with plants. [3,4]) Unfortunately, it’s highly unstable at 37? and requires a tRNA scaffold to function in vivo. [1,7]
Combining SELEX with FACS using a derivative of Spinach, Spinach2, as the selection starting point enabled the selection of a new LURA with better affinity to the ligands. The 49-nucleotide-long RNA oligo named Broccoli activates the fluorescence of DFHBI or DFHBI-1T. It also has improved intracellular stability. [8]
Taking the fruit and veg theme and running with it, the 39-nucleotide-long aptamer that binds acetylated triazole orange got the name “Mango.” Mango is a next-generation aptamer with fluorescence that lasts hours rather than seconds. It binds derivatives of trizole orange-acetylated (TO1). If it is biotinylated in the second step, it’s called TO1-Biotin. [1]
As of 2021, tens of aptamers with fluorophore colors running from yellow to blue are available, and their number keeps growing (Table 1).
Name | Fluorophore | Kd (nM) | Brightness* |
Spinach | DFHBI | 540 | 17.5 |
Spinach2 | DFHBI-1T, DFHBI-2T | 560 | 29.1 |
Broccoli | DFHBI-1T | 340 | 27.8 |
Mango | TO-1, TO-3 | 3 | 10.85 |
Red broccoli | DFHO | 206 | 11.9 |
Corn | DFHO | 70 | 7.25 |
MG aptamer | Malachite Green | 117 | 28.0 |
SRB aptamer | Patent Blue | 23 | N/A |
Table 1. The most commonly used light-up aptamers (table adapted from Bouhedda et al. [9]; see more examples and primary references therein). *Brightness calculated as = (? × ?complex)/1000
As you can see from the table, the first-generation “vegetable” aptamers are not very bright, usually less than a medium-level expressed GFP. But never fear, there’s a new crop coming to overcome this problem. The “pepper” aptamer was initially developed to bind an unstable fluorescent peptide. The second generation of Peppers binds eight fluorescent dyes, ranging from cyan to red, and a few can be co-expressed. [10]
How Can I Get Light-up RNA Aptamers?
If an existing aptamer can help you answer your research question, you’re in luck. If you want to apply LURAs to a new research question, this is a task that can best be likened to obtaining a primary antibody for the protein of your choice.
You can either develop a required LURA yourself, adding a valuable skill to your CV, or buy it, saving time and probably improving your chances of success. It’s too niche and cutting edge a technology for the large biotech companies to be interested in, but smaller ones will help you.
To hear more about RNA aptamers and their development and to understand why they are primarily named after vegetables, listen to The Microscopists Podcast with Rita Strack, who was involved in the development of fluorescent proteins and RNA aptamers (including Spinach2).
Do you use LURAs in your research? Let us know how in the comments below!
References
- Ouellet J. (2016) RNA Fluorescence with Light-Up Aptamers. Front Chem. 4:29.
- Sando, S., Narita, A., Hayami, M., and Aoyama, Y. (2008). Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: a blue fluorescent RNA. Chem. Commun. Camb. Engl. 33:3858-60.
- Paige, J. S., Nguyen-Duc, T., Song, W., and Jaffrey, S. R. (2012). Fluorescence imaging of cellular metabolites with RNA. Science 335(6073):1194
- Paige, J. S., Wu, K. Y., and Jaffrey, S. R. (2011). RNA mimics of green fluorescent protein. Science 333:642–646.
- Strack RL, Disney MD, Jaffrey SR. (2013) A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 10(12):1219-1224.
- Nilaratanakul V, Hauer DA, Griffin DE. (2017) Development and characterization of Sindbis virus with encoded fluorescent RNA aptamer Spinach2 for imaging of replication and immune-mediated changes in intracellular viral RNA. J Gen Virol. 98(5):992-1003.
- A. Weinstein (2019). The Fluorescent Vegetables in Aptamer Soup. Blog.addgene.org
- Filonov, G. S., Moon, J. D., Svensen, N., and Jaffrey, S. R. (2014). Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 136:16299–308.
- Bouhedda F, Autour A, Ryckelynck M. (2017) Light-Up RNA Aptamers and Their Cognate Fluorogens: From Their Development to Their Applications. Int J Mol Sci.19(1):44.
- Strack, R. (2019) A peck of Peppers. Nat Methods 16:1075.