You spent the last few weeks tweaking your Co-immunoprecipitation conditions, testing different antibody/bead combinations, and sampling a panaply of solutions and FINALLY! You have your Co-immunoprecipitation (Co-IP) elution…
Now what?
Well, you have a few choices. It really all depends on what you need know about the proteins in your elution. Do you need to identify the proteins in your elution? Or merely confirm their presence? Do you need to know how the proteins have changed with a treatment? Read below to learn what you should do:
Step 1. Divide and Conquer
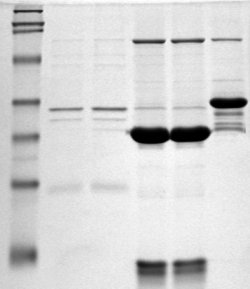
Most protein detection and identification techniques first require you to isolate your proteins. The most common way to do this is to separate your elution proteins by SDS-Polyacrylamide Gel Electrophoresis, SDS-PAGE. SDS-PAGE uses two thin gels, a stacking and a resolution gel, to separate proteins based on size and charge. The stacking gel ensures all proteins, regardless of size or charge, enter the gel at the same time. The resolving gel gently uses electrical currents to pull the proteins through the gel, where small proteins travel faster than larger proteins resulting in separation.
Step 2. Give it the Silver Treatment…or Blue, or Ruby…
Next you need to visualize the mystery proteins in your SDS-PAGE by staining. Since SDS-PAGE separates your proteins by size and charge, staining will give you a quick picture of what proteins of what size you pulled down in your elution. To do to this staining, as with most lab technique, you have several protocols to choose from:
Enjoying this article? Get hard-won lab wisdom like this delivered to your inbox 3x a week.

Join over 65,000 fellow researchers saving time, reducing stress, and seeing their experiments succeed. Unsubscribe anytime.
Next issue goes out tomorrow; don’t miss it.
Coomassie blue staining
Coomassie blue staining is a quick and easy way to peek at the proteins you pulled down. Generally, each band on your Coomassie stained SDS-PAGE represents one protein, the density of which indicates that protein’s abundance. Although Coomassie blue is not the most sensitive staining method, it is advantageous because it leaves your stained proteins suitable for other downstream applications, such as Western blotting or mass spectrometry.
Silver Staining
Silver staining of SDS-PAGE is a more sensitive protein detection method than Coomassie staining. Additionally, the granularity of silver staining makes identifying and isolating protein bands from your SDS-PAGE easy. However, silver stain reagents are more expensive than Coomassie blue reagents and a silver staining protocol takes more supervision. Additionally, silver staining protocols use formaldehyde that can alter Lysine residues and lead to the mis-characterization of Lysine-rich proteins in downstream mass spectrometry experiments.
Ruby Staining
Fluorescent staining is the most recent advance in SDS-PAGE visualization. Fluorescent staining uses dyes with simple binding interactions, rather than covalent reactions (like in silver staining) to mark your proteins. This means that fluorescent staining is less damaging to your proteins’ chemistry. A popular fluorescent stain is SYPRO Ruby Protein Gel Stain. This particular stain is both highly sensitive and compatible with most downstream proteomic applications and mass-spectrometry. However, Ruby staining is pricey and requires the use of a UV-light box, which can be inconvenient.
Step 3. Positively Identify Your Proteins
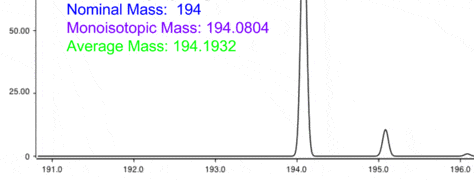
By Mass Spec
If your quest includes identifying the mystery proteins complexing with your protein-of-interest, mass spectrometry (or “mass spec” for those in the know) might be your next step. Mass spec will be covered in more detail in future articles, but in brief: Mass spec is a technique that uses high energy to ionize proteins and separate them on a mass-to-charge ratio. This mass-to-charge profile is then used to positively identify any pulled-down proteins.
Of course, the equipment needed for mass spec experiments are expensive and delicate. Therefore, few labs will buy their own mass spec. Instead, check with the core laboratories on your campus to see what is available to use. Alternatively, there are many commercial mass spec companies that you can hire to identify the proteins in your elution or excised SDS-PAGE bands.
Or By Going West!
If you think you know the identity of the proteins complexed with your protein-of-interest, you may skip the mass spec and go straight from your SDS-PAGE to Western bloting. In Western bloting, your SDS-PAGE separated proteins are transferred with electric current to a membrane, which is then interrogated by antibodies. These antibodies allow you to visualize, positively identify and approximate the interrogated protein’s abundance.
Western blots are great because they can provide you with more information than just “what protein is present”. Depending on what type of antibodies you use for interrogation, Western bloting can also tell you about the post-translational modifications found on your pulled-down proteins, such as phosphorylation, sumoylation, or ubiquitination, allowing you to learn about how your mystery protein complex is modified.
So, are you ready to find out what mysterious protein is dancing with your protein-of-interest?
You made it to the end—nice work! If you’re the kind of scientist who likes figuring things out without wasting half a day on trial and error, you’ll love our newsletter. Get 3 quick reads a week, packed with hard-won lab wisdom. Join FREE here.