If you are working in the biomedical research field and you haven’t yet heard about CRISPR Cas9 gene editing technology, you have some catching up to do – but don’t worry, this article will bring you up to speed about CRISPR in no time!
In the few years since the CRISPR phenomenon emerged, this technology has captivated researchers spanning the entire biomedical field. This is not only because of its powerful gene-editing capability but also because of its accompanying ethical considerations.
First, let’s briefly touch upon the history that gave rise to the CRISPR genome editing technology. Then, the key players in the system will be described, and we will examine how scientists adopted and continue to adopt this system for genetic engineering.
An Ancient Defense System – CRISPR Explained
Surprisingly, it turns out that an amazing defense system sits in the genome of many types of bacteria. CRISPR stands for “clustered regularly interspaced short palindromic repeats.” For a long time, bacteriologists realized that many bacterial genomes contained a region defined by a variety of unique sequences called “spacers” that are flanked by short inverted palindromic repeats. At first, this finding puzzled bacteriologists, until someone decided to align those unique sequences to known DNA libraries. The result they got was astounding – a surprisingly large number of those unique sequences matched to bacteriophage DNA sequences! That’s when researchers realized that they might be looking at a bacterial immune system, used to defend against bacteria’s natural predator, the bacteriophage.
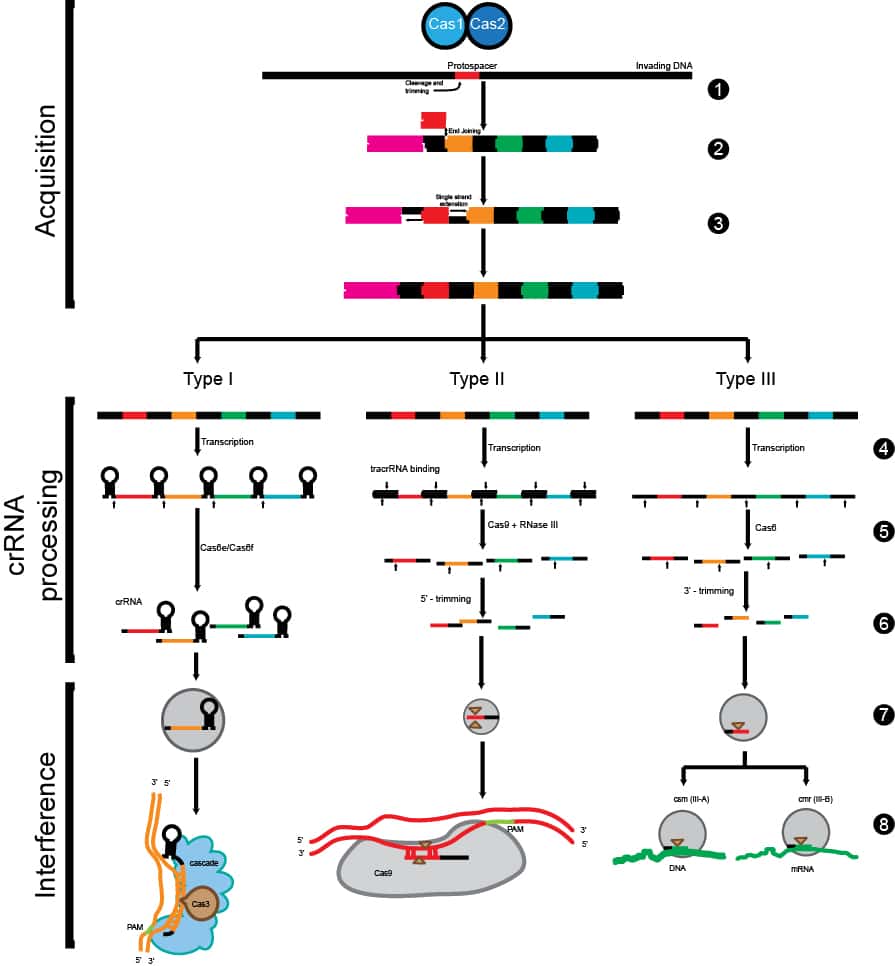
Once researchers set their sight in the right direction, they were able to map out and characterize the major components of the system. So far, three different CRISPR pathways have been mapped out, and the CRISPR II system seems to be the most prominent (Figure 1).
The CRISPR system establishes immunity by “remembering” components of the invading pathogen (i.e., bacteriophages) through storing the unique bacteriophage DNA sequences between the palindromic repeats. The next time the same bad guys try to invade the bacteria, the CRISPR system and a key enzyme called CRISPR-associated protein 9 (Cas9) scan the DNA pool via the use of complementary guide RNAs (gRNAs) for any intruder that matches the existing unique sequences stored in the “databank.” Cas9 then nicks the DNA and signals the DNA demolition team downstream to destroy the invader. How neat is that?!
Making Gene Editing Easier
Before CRISPR genome editing technology emerged, scientists relied on techniques such as homologous recombination, zinc finger nucleases (ZFNs) and transcription activator like-effector nucleases (TALENs) for gene editing. Compared to the ZFNs1 and TALENs, the CRISPR system is highly specific, cost-effective, and much easier to design and use. Another major advantage of CRISPR is the capability to “scale up.” Multiple gRNAs can be used to edit different genetic loci simultaneously, allowing you to study diseases that involve multiple genes.
Hopefully by now you can see the great potential of CRISPR technology to generate any gene knockout of interest. All you have to do is design a gRNA sequence,2 and co-transfect an expression plasmid containing Cas9 into you cells of your interest (that’s right, CRISPR works in many different cell types!). With a good setup, you are almost guaranteed to get the desired mutation following some screening. In addition, scientists have also modified the Cas9 enzyme to further enhance the specificity of the knockout region.3
CRISPR Technology in Action
Scientists are already testing the power of CRISPR technology, as witnessed by the surge in CRISPR technology-related publications. Some of the most notable examples to date are:
Infectious Diseases and Allergy
HIV – The human immunodeficiency virus (HIV) integrates into our genome and causes AIDS. Consequently, scientists are contemplating how to use CRISPR to either inactivate or remove the integrated HIV genomes in HIV-infected patients. Scientists have already demonstrated that CRISPR-technology can remove the integrated HIV DNA in vitro. The verdict is still out to see if this technology will work in the clinical setting.4 For a recent review article on the status of CRISPR in the field of immunology and allergy, look here.
Allergy – Allergy is another area that is receiving considerable attention among CRISPR enthusiasts. While allergy itself is generally well managed by affected individuals, being allergic to eggs means that 2% of children worldwide cannot avail of many of the routine vaccinations that are produced using chicken eggs. Given that most egg allergies are believed to be related to just four egg-white proteins, researchers in this area believe that they can use CRISPR to produce hypoallergenic eggs. Expression of mutated egg-white proteins in bacteria has already resulted in proteins that don’t trigger a reaction in the serum of allergic individuals, and the first hypoallergenic chickens are already in the pipeline!
Genetic Diseases
Sickle-Cell Anemia – A team of researchers from Boston used CRISPR technology to edit a stretch of DNA that constitutes the “enhancers” in the blood stem cells. By disabling the enhancers, scientists saw a dramatic increase in the fetal hemoglobin – a big step towards gene therapy for sickle cell diseases.
Cystic Fibrosis – Scientists have replaced the defective CFTR gene with the wild-type gene using the CRISPR system. And the functional CFTR gene restored function in vitro.5
Generation of Knockout Animals
Using CRISPR, it is now possible to generate knockout animals with high specificity and efficiency because both copies of the target can be removed simultaneously. As a result, knockouts can be generated in shorter timeframes, as CRISPR begins to replace traditional knockout protocols, which typically require further screening and breeding of the animals.6,7 Check out our related article to find out how to get started using CRISPR Cas9 in Drosophila
Genetic Screening for Loss of Function
You can also use a CRISPR screen for loss-of-function mutations using an assortment of RNA sequences (an RNA library).8
Nature special: CRISPR
Cancer Therapy
The development of cancer therapy using gene editing has become one of the lastest focuses of CRISPR technology. By editing T-cells to remove genes that can allow cells to be inactivated by cancer cells, adding genes that allow immune cells to better target cancers or a combination of both approaches, researchers provide a way to harness the body’s own defense mechanisms to better treat various cancers. [9]
Parting Words
So that was a brief introduction to CRISPR technology, which is hailed as the next “breakthrough” in gene editing (check the main pros and cons of CRISPR-Cas9). Don’t forget that the key to its specificity lies in the design of the gRNA sequence, so before you jump on the bandwagon, make sure you consult some references on designing gRNAs.
Finally, and as we inch towards personalized gene editing and medicine, we should always keep in mind the kind of genetically modified organisms we might be creating and the potential biological and ethical consequences. Otherwise, enjoy the technology and let us know what you think by writing in the comments section!
Discover more about CRISPR in the Bitesize Bio CRISPR Research Hub.
And because there’s so much to learn, get your free gene editing 101 eBook to get up to speed today!
References
- Urnov FD, et.al. (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 435: 646–51. doi:10.1038/nature03556.
- Fu Y, et. al. (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology. 32: 279–84. doi:10.1038/nbt.2808.
- Mali P, et al. (2013) RNA-guided human genome engineering via Cas9. Science. 339: 823–26. doi:10.1126/science.1232033.
- Ebina H, et. al. (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Scientific Reports. 3. doi:10.1038/srep02510.
- Schwank G, et al. (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13: 653–58. doi:10.1016/j.stem.2013.11.002.
- Hai T, et. al. (2014) One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Research 24: 372–75. doi:10.1038/cr.2014.11.
- Gratz S, et. al. (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–71. doi:10.1534/genetics.113.160713.
- Koike-Yusa H, et.al . (2013) Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature Biotechnology 32: 267–73. doi:10.1038/nbt.2800.
- Zhang X, et al. Engineering T Cells Using CRISPR/Cas9 for Cancer Therapy. Methods Mol Biol. 2020;2115:419-433. doi:10.1007/978-1-0716-0290-4_23
Originally published in January 2017. Updated and republished in April 2017.