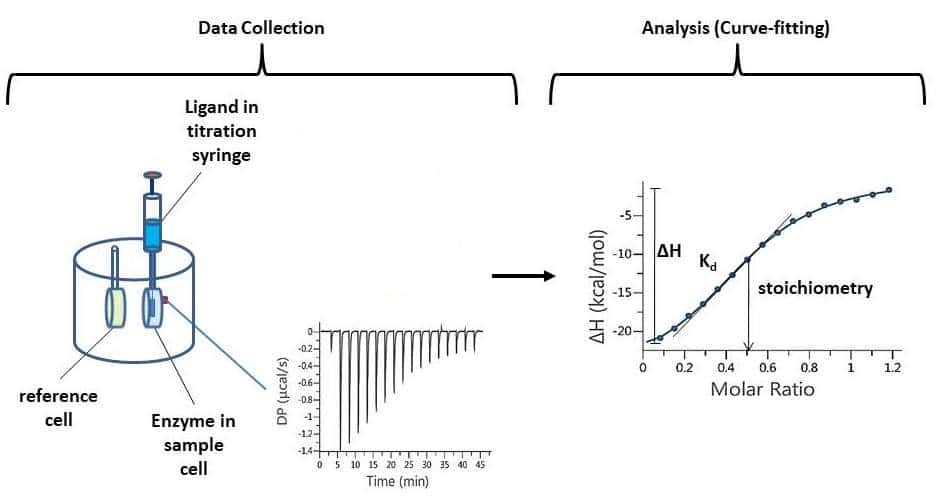
What is Isothermal Titration Calorimetry?
Isothermal titration calorimetry (ITC) measures the heat generated (or absorbed) when one solution is titrated into another. Most commonly, a small molecule or peptide is titrated into a protein. If the molecule binds to the protein, heat is given off (or absorbed) with each injection, until the protein is saturated.1 This binding curve (Fig. 1) can then be analyzed to find the dissociation constant (Kd), molar free energy change (ΔG), molar enthalpy change (ΔH), molar entropy change (ΔS) and stoichiometry of the protein-ligand binding interaction.
The whole process is fairly hands-off: the ‘cell’, which contains the protein is loaded by hand with a Hamilton syringe; the titration syringe is loaded from a PCR tube with the click of a button; and automated control software takes care of the rest. A typical experiment takes 2 – 3 hours (from initial setup to output Kd).
What is Isothermal Titration Calorimetry Used For?
There are loads of ways you can use ITC. You can test how mutations in a protein affect ligand binding to find possible molecular rationale for the effects of disease mutations. You can derive structure affinity relationships for small molecules (or peptides or DNA!) and their target enzymes. You can use competition experiments to see if inhibitors compete with a particular substrate. All these rely on the Kd values, but the stoichiometry is also useful: comparing the theoretical and experimental stoichiometry values of a ligand implies what percentage of a protein is correctly folded. These are some common uses but by no means an exhaustive list!
What’s the Benefit of Using Isothermal Titration Calorimetry over Other Techniques?
The unique selling point of ITC is that no labelling or immobilization of the binding partners is needed [unlike techniques such as surface plasmon resonance (SPR), enzyme-linked immunosorbent assay (ELISA), etc.]. This means there’s no need to worry about tags or immobilization surfaces obscuring biologically relevant interactions.
Secondly, affinities from nanomolar through to millimolar can (in theory) be quantified. Modifying the experiment to accommodate different affinities is generally straightforward, though there are practical limitations such a solubility (an issue for weak interactions) and signal-to-noise (an issue when determining the Kd for strong interactions). This second limitation may sound counterintuitive, but is a consequence of needing a sigmoidal titration curve. The range of ligand affinities that are measurable for a particular protein can be widened by using competition experiments.
A wide range of buffers are tolerated (though some are better than others). This might become important if your protein needs a particular pH or additive to stop it precipitating, or if you want to test how changes in pH, ionic strength or metal ions (to name a few) affect a binding interaction. Since no special additives or reagents are required, the cost of running an experiment is very low – all that is needed is the ligand and receptor in whichever buffer keeps them soluble and stable.2
Another great thing about ITC is that the Kd, ΔG, ΔH, ΔS and stoichiometry can be determined in a single experiment.3 That’s a ton of information from a few hours of lab work. It can even be possible to extract multiple binding constants (if the protein has two ligand binding sites with different Kds) from a single (well-planned) titration.
ITC can do kinetics, too. Enzyme reaction kinetics and ligand binding kinetics can both be studied. These methods can be used to find steady state enzyme kinetic parameters and rate constants for ligand binding.
Disadvantages of Isothermal Titration Calorimetry
There are, of course, situations where alternative techniques are more appropriate.
The first key drawback of ITC is its low-throughput nature. Only one ligand can be tested at a time, and (as mentioned above) each titration takes (at least) 3 hours. Automated systems exist (see here and here), but given the finicky nature of cell loading, I would personally be quite reluctant to leave my precious protein sample in the hands of an automated system!
The second major issue with ITC is that it uses a lot of protein compared to lower-volume techniques such as fluorescence polarization, thermal shift and ELISA.
For both of these reasons, ITC is obviously not suited to screening large numbers of ligands (e.g. in the context of drug discovery). It is also not feasible if protein or ligand availability is very limited (see below).
Sounds Great! Can I Try with My Protein/Ligand?
In short, (probably) yes!
ITC can, in principle, be used to characterize any interaction or process that involves the generation or absorption of heat.
I’d guess that the most difficult step for most people would be finding a calorimeter they can use (they’re expensive and delicate, so you’ll probably have to ask nicely!). If you’re lucky enough to have access to one, you’ll likely need more protein than other binding assays. The exact amount will vary greatly depending on the Kd and ΔH of the interaction (see Table 1 for a rough idea). If you have no idea how strong your binding interaction is, then a commonly recommended starting point is 30 µM cell concentration and 300 µM syringe concentration. For a 30 kDa protein and a cell volume of 200 µL (which is standard for Malvern’s Peaq and ITC200 models and TA instrument’s Low Volume Nano ITC) this means that 270 µg protein is needed.4 The ligand volume needed for these instruments is 80 µL, so 0.024 µmol ligand would be needed for the ligand-to-protein titration, and a further 0.024 µmol for a ligand-to-buffer control experiment.
Anticipated Kd | Recommended protein in cell | Mass of protein needed (assuming MW = 30 KDa and 300 µL needed to fill cell) |
0.1 µM | 1 µM | 9 µg |
1 µM | 10 µM | 90 µg |
10 µM | 100 µM | 900 µg |
There’s plenty of information online about experimental design, troubleshooting, and overcoming known experimental limitations (for total ITC beginners, I’d recommend these slides written by Peter Gimeson from Malvern. The manuals that come with each machine are also really helpful).
Footnotes
1Of course, there are a long list of reasons that this may not happen!
2DISCLAIMER: This is true, but more commonly a few ‘trial’ titrations are needed to figure out the best conditions.
3The titration cell is made of Hastelloy®. Some useful information on recommended buffers can be found here.
4For a cell volume of 200 µL, ~300 µL is needed to fill the cell due to dead volume in the Hamilton syringe and the stem of the sample cell.