What is the first image which comes to mind when you think about microalgae? Green scum that covers the surfaces of ponds? Unsightly stains on pavements and walls? Far from being a nuisance in ponds, lakes, drains and on surfaces, microalgae are fascinating microorganisms which are used to understand various biological processes. Microalgae have been used as model microorganisms to study several mechanisms, including membrane permeability, osmoregulation, salt tolerance, and the transition to multicellularity. Microalgae have also been used as feedstocks in a number of applications such as biofuel, bio-fertilizer, animal feed, and pharmaceuticals.
Laboratory techniques such as culture and DNA extraction have been developed for microalgae. Genetic transformation of microalgae paves the way for genetic engineering of these microorganisms, enabling us to explore the potential of microalgae fully. This article will introduce some of the common ways to introduce DNA into microalgae.
Note: Like most microorganisms, transformed microalgal cells can be differentiated from non-transformed microalgae using selection methods such as antibiotic resistance and metabolic markers. Remember to include a selectable marker together with the gene(s) to be transformed.
Particle Bombardment (Gene Gun)
A popular method of introducing exogenous DNA into microalgal cells is particle bombardment. During particle bombardment, tungsten or gold microprojectiles are coated with DNA and “fired” at microalgal cells using a gene gun – a particle delivery system which accelerates microprojectiles with a burst of pressurized gas. The DNA carrying particles penetrate the cytosol and nucleus, thus delivering the transgenes to the cells. The advantage of using particle bombardment to transform microalgae is its high success rate. Particle bombardment has been successfully used to transform a variety of microalgal species, with and without cell walls (Doron et al, 2016). Particle bombardment can also be used to transform cell organelles such as chloroplasts and mitochondria.
Inert Particle Mediated Transformation
A method of microalgal transformation which does not require the use of complicated equipment is the use of inert particles to deliver foreign DNA into cells. In this method, macro or microparticles (e.g. glass beads or silicon carbide whiskers), the gene(s) of interest to be delivered, denatured fish sperm DNA which binds to nucleases in the cell and prevents degradation of the gene(s) of interest, as well as polyethylene glycol to increase transformation efficiency are added to microalgal cells, and the suspension is agitated by vortexing. Microalgae with and without cell walls have been successfully transformed using this method (Doron et al, 2016). Higher transformation efficiencies are obtained with microalgae with thin or no cell walls. Some microalgae like Dunaliella sp. naturally do not have cell walls and can be directly transformed by inert particle mediated transformation. For microalgae with cell walls, cell wall deficient mutant strains of the microalgae are recommended for maximum efficiency of transformation using this method. Alternatively, cell walls of microalgae can be digested with enzymes such as subtilisin and lysozyme to remove cell walls before transformation. After transformation, microalgal cells are kept in isotonic solutions of mannitol or sorbitol for cell wall regeneration to occur. (Coll, 2006).
Electroporation
Electroporation can also be used to transform microalgae with reduced cell walls. Microalgal cell suspension together with DNA is placed in an electrocuvette and an electrical field is applied using an electroporator. The electric pulse causes the formation of pores in the plasma membrane, thus allowing the DNA to enter the cell. Electroporation is a rapid process which allows microalgae to be transformed quickly. By varying electroporation settings such as voltage, a variety of microalgae, such as Chlamydomonas, Chlorella, Neochloris, and Acutodesmus have been transformed (Muñoz et al, 2018).
Agrobacterium Mediated Transformation
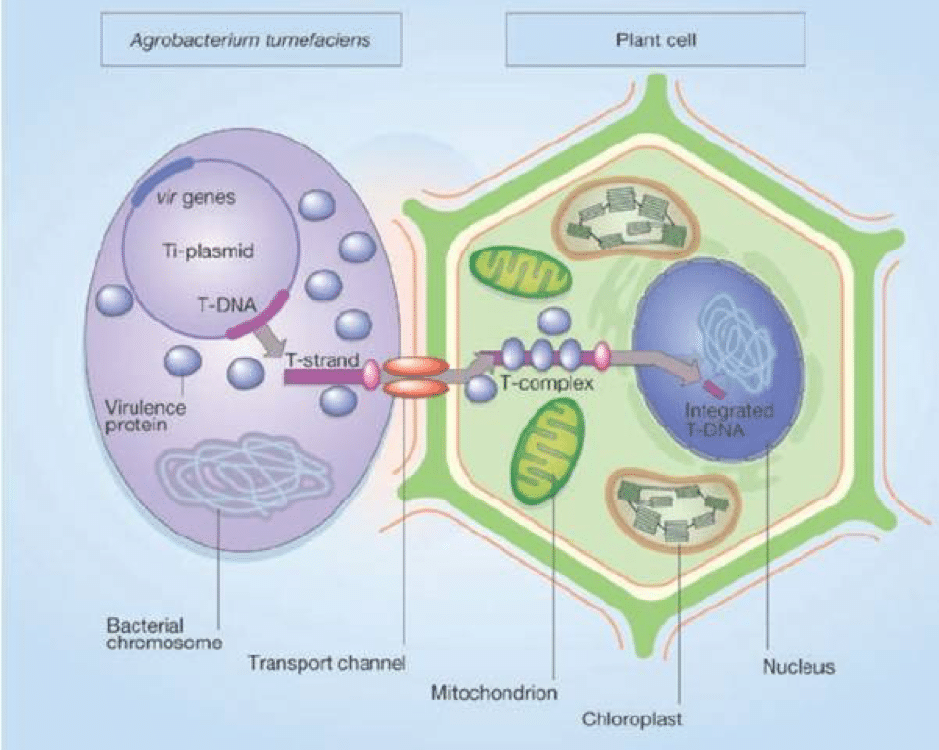
The bacterium Agrobacterium tumefaciens is a plant pathogen which has been widely used in the genetic engineering of plants due to its ability to transfer DNA to the plant host (Gelvin, 2003). Microalgae can also be transformed by Agrobacterium. When Agrobacterium infects a plant, it transfers exogenous T-DNA from the bacterial tumour-inducing (Ti) plasmid to the plant cells. The T-DNA integrates into the genome of the plant and expression of tumour-inducing genes on the T-DNA leads to the formation of tumours (Figure 1).
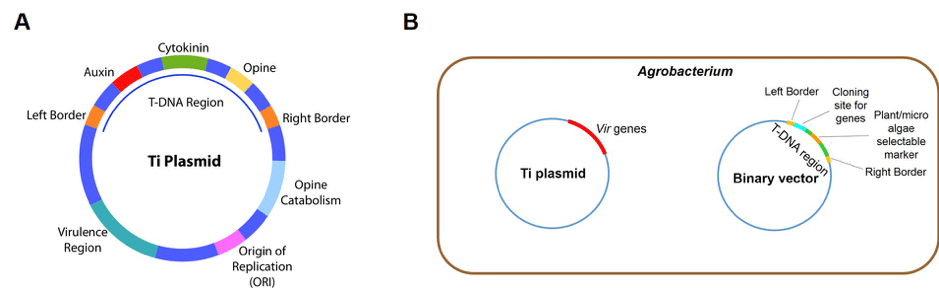
For efficient transformation of plants and microalgae in the laboratory, a binary vector system based on the principles of T-DNA transfer is used in Agrobacterium mediated transformation. The T-DNA region is located on a T-DNA binary vector while the Vir genes which express virulence proteins required for T-DNA transfer are contained in a separate Ti plasmid (Figure 2). The foreign genes, together with a plant or microalgae selectable marker, are cloned into the T-DNA region flanked by the border repeats and the Agrobacterium is induced to infect plant tissue and microalgal cells. Transformed plants and microalgae are selected by antibiotic resistance. Only those containing the selectable marker and hence, the transgenes, will grow on culture media containing the antibiotic.
These transformation techniques enable us to better understand microalgae and manipulate them to greater extents. Who knows what else we can learn from these weird and wonderful microorganisms in the years to come?
References
- Coll J.M., 2006. Review. Methodologies for transferring DNA into eukaryotic microalgae. Spanish Journal of Agricultural Research 4(4): 316-330.
- Doron L., Segal N. and Shapira M., 2016. Transgene Expression in Microalgae—From Tools to Applications. Front. Plant Sci. 7: 505. doi: 10.3389/fpls.2016.00505
- Gelvin S.B., 2003. Agrobacterium-Mediated Plant Transformation: the Biology behind the “Gene-Jockeying” Tool. Microbiology and Molecular Biology Reviews 67(1): 16-37.
- Muñoz C.F., de Jaeger L., Sturme M.H.J., Lip K.Y.F., Olijslager J.W.J., Springer J., Wolbert E.J.H., Martens D.E., Eggink G., Weusthuis R.A., Wijffels R.H., 2018. Improved DNA/protein delivery in microalgae – A simple and reliable method for the prediction of optimal electroporation settings. Algal Research 3: 448-455.