Why Study Mitochondrial DNA?
Mitochondria produce the majority of adenosine triphosphate (ATP) that is required to power a multitude of cellular processes. These cellular powerplants have a 16-kb circular genome that encodes a handful of proteins, [1] and mutations in mitochondrial DNA (mtDNA) are associated with a wide range of diseases, including hearing loss, muscle disorders, diabetes, neurodegenerative disease, and cancer. [2,3]
Before you can sequence mtDNA to study the effect of these mutations in these and other disorders, you need first to isolate it. mtDNA differs from nuclear DNA in several essential ways. To ensure your results are reliable when isolating and sequencing mtDNA, you need to understand and take into account these differences.
Features of mtDNA
Hundreds of Copies per cell
Most eukaryotic cells contain hundreds of mitochondria, with hundreds of copies of mtDNA (estimated 2-10,000 mtDNA copies per cell).[4] Because of this, it is common for individual mutations to be present in only a subset of mitochondria. Thus, not all of these mitochondrial genomes are identical within a cell or organism. This is referred to as heteroplasmy.
High mutation rate
In most organisms, mitochondria are maternally inherited, and because of this uniparental inheritance recombination is limited. Other causes of variation in the mitochondrial genome include the lack of comprehensive DNA repair mechanisms [5] and histones, and a highly oxidative environment; all of which contribute to increased mutation rates.
This heteroplasmy can make identifying disease-associated mutations more challenging since mutations in mtDNA often require a threshold level to be reached before clinical symptoms are observed. [6] Techniques for isolating mtDNA need to retain this heteroplasmy and allow for quantification.
Microheteroplasmy
The mitochondrial genomes of one organism can house hundreds of independent mutations that are found in 1-2% of said mitochondrial genomes. This microheteroplasmy further complicates the analysis of mtDNA, as these mutations are not quantifiable by standard PCR techniques.
Existence of Nuclear Mitochondrial Pseudogenes
Finally, nuclear mitochondrial (nuMT) pseudogenes arise from the integration of mitochondrial genes into the nuclear genome, usually in non-coding regions. The existence of nuMTs is a confounding factor in the identification of mitochondrial genetic variants. [7] mtDNA extraction methods should effectively remove nuclear DNA to limit the interference of nuMTs in downstream analyses.
Despite the high copy number, mtDNA contributes only 0.2% of the total DNA content of a cell. This minority status, along with the observed heteroplasmy of mtDNA, requires samples to be enriched for mitochondrial DNA before sequencing to maximize the ability to detect mutations in mtDNA.
How to Enrich for Mitochondrial DNA
There are various methods available for enriching mtDNA, but before choosing one, you should consider the following:
- Purity and enrichment levels you require for your downstream analysis
- The time you have available to spend on the enrichment process
- Level of skill and mastery required to perform the method.
Density Gradient Centrifugation For Isolating Mitochondrial DNA
If you have access to an ultracentrifuge, the gold standard of mtDNA enrichment is through density gradient centrifugation. In this method, total DNA is loaded onto a cesium chloride density gradient and centrifuged for 10 hours at 450,000 x g to separate the DNA by size. The result of this ultracentrifugation is two distinct bands of DNA; a higher nuclear DNA band and a lower mitochondrial DNA band.
Addition of ethidium bromide enhances the density differential, permitting better visualization of the DNA and aids in the collection, which is achieved by inserting a needle into the tube at the appropriate location and removing the lower band enriched in mitochondrial DNA.
While this is an effective method for isolating mtDNA, there are several drawbacks, including:
- A long ultracentrifugation step
- The use of dangerous and toxic chemicals
- Patience and dexterity to collect the mtDNA band.
Also, the force generated during the ultracentrifugation step and the use of hypodermic needles to extract the DNA from the ultracentrifuge tube may result in DNA shearing.
PCR Using Mitochondrial Genome-Specific Primers
Another low-cost method available to enrich for mtDNA is to perform PCR using mtDNA-specific primers. PCR enrichment for mtDNA can be achieved either through using a collection of primers to amplify the mtDNA in fragments or by long-range PCR using just one or two primer sets. [8,9]
Manual protocols have been optimized that combine conventional miniprep kit, paramagnetic bead-based purification, and limited PCR amplification to enrich mtDNA. This method has the added benefit that it does not require any specialized equipment, only access to a PCR machine.
While PCR amplification does offer a cost-effective and efficient way to enrich for mtDNA, the method suffers from some limitations.
- The polymerase used can introduce errors, making it difficult to distinguish true mutations in the mitochondrial genome.
- PCR-based methods are also unsuitable if you wish to interrogate the mitochondrial epigenome as epigenetic markers are lost or altered during the amplification process.
Mitochondrial Isolation and Enzymatic Enrichment
Another approach is to separate and isolate the entire mitochondria organelle before purifying the mtDNA. There are several methods available, including differential centrifugation, which uses several rounds of gradient centrifugations to isolate intact mitochondria from a cellular lysate.
Alternatively, some commercially available kits use antibodies for TOM22, a core component of the mitochondria outer membrane protein translocation pore, bound to magnetic beads to isolate the mitochondria. [10]
Enrichment of mtDNA via exonuclease digestion increases the success of both of these methods above significantly. Since mitochondrial DNA is circular and nuclear DNA is linear, an exonuclease that digests linear DNA provides further enrichment of mtDNA. [11]
While combining mitochondrial isolation and exonuclease digestion is a successful way to enrich for mtDNA, this method has the following restrictions:
- It is a time-consuming, multistep approach
- Uses multiple rounds of centrifugation which may damage mtDNA and lead to contamination with nuclear DNA (if isolating mitochondria by differential centrifugation)
- Requires specialist equipment in the form of a magnet to capture the mitochondria bound to the beads (if using magnetic beads to capture mitochondria).
A Simpler Approach to Enrich For Mitochondrial DNA
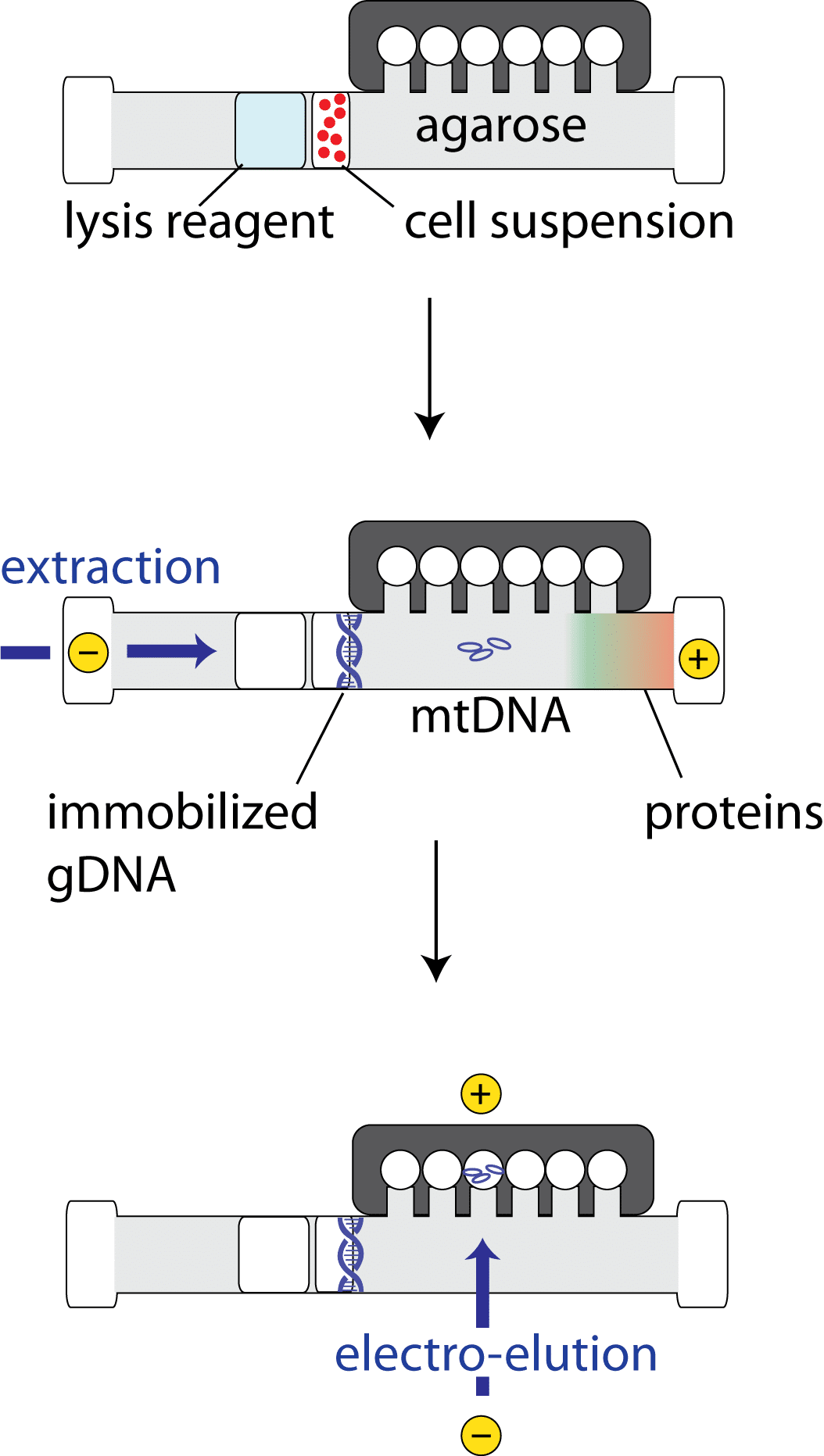
The SageHLS™ system from Sage Science offers a more straightforward and automated method for the enrichment of mtDNA. In this system, 1-1.5 million whole cells are loaded onto a proprietary agarose gel cassette alongside lysis buffer, which is transferred into the sample well by electrophoresis. Once cell lysis has occurred, the cell contents move through the agarose by electrophoresis.
Cellular material, including proteins, RNA, and lipids, move rapidly through the agarose. Nuclear DNA, which remains mostly of chromosomal length, is retained in the sample well, while the smaller mtDNA migrates into the gel, allowing for their separation. Electro-elution is then used to recover the mtDNA which is ready for use in downstream applications such as sequencing on the Illumina® platform. [12]
SageHLS offers significant benefits for the enrichment of mtDNA.
- Simple. No prior lysis of cells required.
- Quick. Enrichment of mtDNA from whole-cell samples takes less than 3 hours.
- Efficient. No centrifugation steps are required, and nuclear DNA remains chromosomal in length, minimizing nuMTs in the sample.
- Automated. After cells are loaded onto the agarose gel, the extraction and size selection is fully automated, meaning you can walk away and focus on other tasks.
- Effective. Purity levels obtained are comparable to density gradient centrifugation, and a 20-fold enrichment of mtDNA has been observed. [12]
- Gentle. DNA is not subjected to lengthy or repeated centrifugation steps, meaning DNA shearing is minimize.
- Results. Isolated mtDNA can be used directly for sequencing on the Illumina platform.
SageHLS can isolate mtDNA from a wide range of samples, including tissues and suspensions cell cultures, as long as you disperse the sample into single cells before loading onto the agarose gel.
mtDNA Extraction Summarized
It is beneficial to perform enrichment of mtDNA before sequencing to minimize the presence of nuMTs which may negatively impact the analysis and results of your research. This enrichment is achievable through multiple methods that take advantage of the differences between nuclear and mtDNA, including size, cellular location, and whether it is linear or circular.
The SageHLS method offers purity and enrichment comparable to the gold standard of density gradient centrifugation in a simple, quick, and automated manner, without the need for ultracentrifugation.
References
- Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. (1981) 9(290):457-65. DOI: 10.1038/290457a0
- Ryzhkova AI, Sazonova MA, Sinyov VV, et al. Mitochondrial diseases caused by mtDNA mutations: a mini-review. Ther Clin Risk Manag. (2018) 14:1933–42. DOI: 10.2147/TCRM.S154863
- Taylor RW & Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. (2005) 6(5):389–402. DOI: 10.1038/nrg1606
- Miller FJ, Rosenfeldt FL, Zhang C, et al. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. (2003) 31(11): e61. DOI: 10.1093/nar/gng060
- Bogenhagen DF. Repair of mtDNA in vertebrates. Am J Hum Genet. 1999 64(5):1276–81. DOI: 10.1086/302392
- Rossignol R; Faustin B; Rocher C, et al. Mitochondrial threshold effects. Biochem J (2003) 370(3): 751–62. DOI: 10.1042/bj20021594
- Simone, D., Calabrese, F.M., Lang, M. et al. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics (2011) 12, 517. DOI: 10.1186/1471-2164-12-517
- Ramos A, Santos C, Alvarez L, et al. Human mitochondrial DNA complete amplification and sequencing: a new validated primer set that prevents nuclear DNA sequences of mitochondrial origin co-amplification. Electrophoresis. (2009) 30(9):1587-93. DOI: 10.1002/elps.200800601.
- Cui H, Li F, Chen D, et al. Comprehensive next-generation sequence analyses of the entire mitochondrial genome reveal new insights into the molecular diagnosis of mitochondrial DNA disorders. Genet Med (2013) 15, 388–94. DOI: 10.1038/gim.2012.144
- Franko A, Baris OR, Bergschneider E, et al. Efficient Isolation of Pure and Functional Mitochondria from Mouse Tissues Using Automated Tissue Disruption and Enrichment with Anti-TOM22 Magnetic Beads. PLOS ONE 8(12): e82392. DOI: 10.1371/journal.pone.0082392
- Gould MP, Bosworth CM, McMahon S, et al. PCR-Free Enrichment of Mitochondrial DNA from Human Blood and Cell Lines for High Quality Next-Generation DNA Sequencing. PLOS ONE (2015) 10(10): e0139253. DOI: 10.1371/journal.pone.0139253
- Boles C & Houde N. Targeted Mitochondrial DNA Extraction and Enrichment Using the SageHLS System. Application Note: SageHLS. Sage Science Ltd. 2019.