Multiplex ligation-dependent probe amplification (MLPA) is a molecular technique developed by MRC-Holland back in 2002. In a nutshell, MLPA is a sensitive technique that allows quantification of nucleic acid sequences, quickly and efficiently. It is performed in many laboratories worldwide, and can be applied to detect copy number changes (like deletions or duplications) of a gene, identify the methylation status of DNA, detect single nucleotide polymorphisms (SNPs) and point mutations, and quantify mRNA. Therefore, it is used in many research and diagnostic fields, such as cytogenetics, cancer research, and human genetics, among others.
How does it work?
MLPA consists of the following steps (Figure 1):
- Denaturation
- Hybridization
- Ligation
- Amplification (by PCR)
- Fragment Separation and Data Analysis
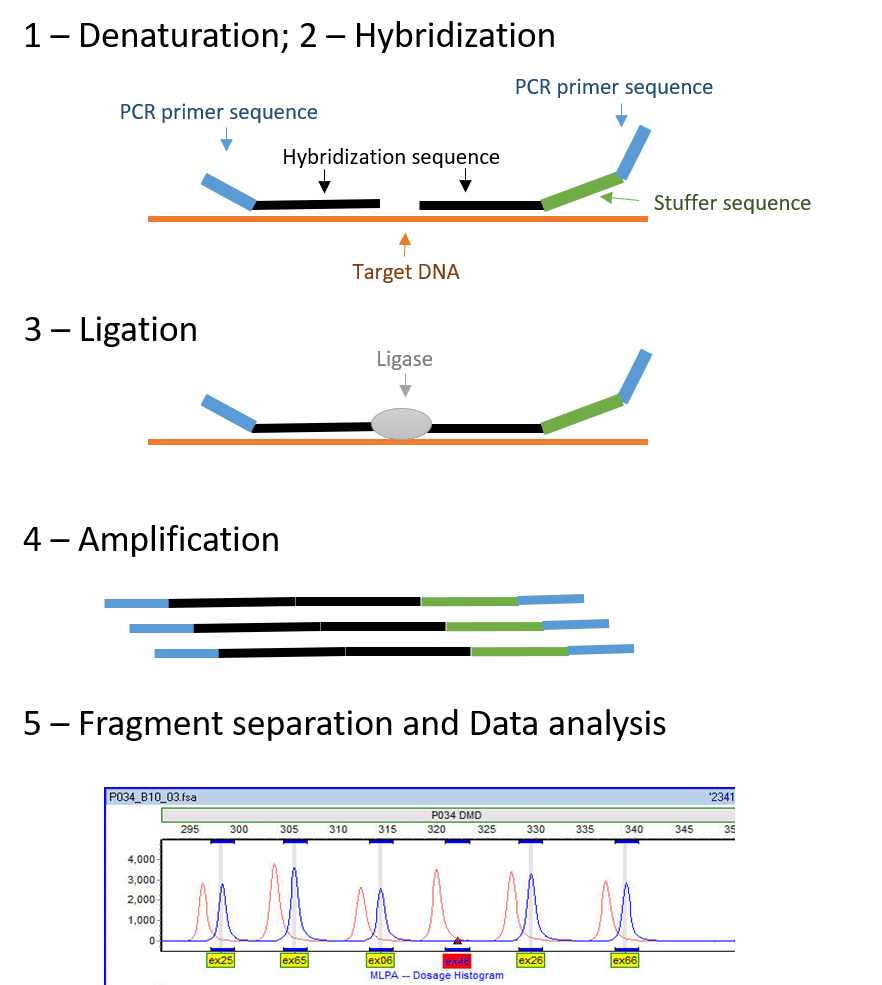
Figure 1 – Visualization of the MLPA technique (adapted from Schouten, Jan P., et al.1). We can also observe a typical electropherogram obtained through MLPA analysis showing a deletion of exon 46 (red arrow). (Electropherogram adapted from https://commons.wikimedia.org/wiki/File:MLPA_in_GeneMarker.jpg)
1-Denaturation and 2 – Hybridization
Denaturation involves separation of the annealed DNA strands, so that double-stranded DNA becomes single-stranded.
Hybridization involves hybridizing the DNA sample to specific probes. Because it is a multiplex technique, you can analyze each sample by up to 60 probes simultaneously, thus targeting different sites!
These probes have a primer sequence that binds to the PCR-primer in the amplification process. All different probes will have the same primer binding sequence. Additionally, the probes also have a hybridization sequence complementary to the target site that will allow the probe to bind to the DNA. Both probes will hybridize on adjacent sites on the DNA strand.
One of the probes from the pair contains a stuffer sequence, which is different in length for each target site. The length of the stuffer sequence changes between different probes, allowing multiplexing. So, you can expect each amplification product to have a unique length!
3-Ligation
The ligation step will bind the two probes together. In this step, a specific enzyme called DNA ligase is used. It binds the probes that are already hybridized on adjacent sites of the DNA strand at the target site. The ligase used in MLPA protocols is ligase-65, an NAD-dependent ligase enzyme, that can also be useful in other applications.
Now, the question stands: if our goal is to ligate both probes, why are they separate molecules to begin with? Well, both probes contain the binding sites for PCR-primers. This means, if we were to use the probes as a single molecule, we would obtain an amplification product, even without the DNA target site, thus giving us non-specific amplification. The enzyme ligase is extremely specific: if there are any mismatches between the probe and the target site, the ligase will not be able to bind the probes and no amplification would occur. Consequently, MLPA detects specific point mutations, and even distinguishes between pseudogenes and the real target gene.
4-Amplification
The next step is amplification, which is essentially a polymerase-chain reaction (PCR) (Table 1). For the PCR step, a polymerase, dNTPs, and a forward and reverse primer are added. Since all of the probes have the same PCR-primer sequence, it will only be necessary to add one pair of universal primers to study all of our targets. The forward primer is fluorescently labelled, allowing visualization and quantitation during analysis.
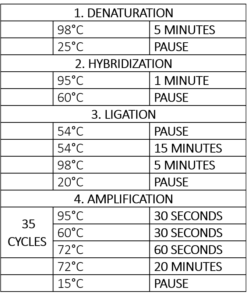
Table 1 – Thermocycler program for the MLPA reaction [adapted from MLPA (Multiplex Ligation-dependent Probe Amplification) General Protocol for the detection and quantification of DNA sequences, from MRC-Holland -https://www.mlpa.com2]
5-Fragment Separation and Data Analysis
After amplification, the fragments are separated by capillary electrophoresis. Capillary electrophoresis separates fragments based on their length, and shows different length fragments as peak patterns, called an electropherogram (Figure 1). Each amplicon has a different known size, due to the stuffer sequence on each specific probe, and therefore each amplicon can be quantified during data analysis.
The data obtained by capillary electrophoresis will be the input for the analysis. MRC- Holland provides a free software for data analysis – Coffalyser.
By comparing each sample to a set of reference samples, we can obtain a probe ratio. This probe ratio will inform us of how many copy numbers a gene has. Since most human genes are diploid, if the sample presents two copies, the ratio will be 1.0; i.e. the sample probes have obtained the same number of genes as the reference sample.
However, if the ratio is 0.5 there was only one copy of the gene in the individual, which probably means a heterozygous deletion of the target gene. If, on the other hand, the ratio is 1.5, there is, probably, a heterozygous duplication of a gene.
MRC-Holland offers many different kits that may have the solution for your problems. However, if you are trying to find something a bit more obscure, or study something that isn’t in any kit, you can design your own probes. I advise you to read carefully the protocol for synthetic probe design.
Advantages of MLPA
- MLPA is a highly sensitive, robust and high throughput technique.
- It can discern between point mutations, as well as duplication/deletion of genes. Therefore, it has a great advantage over other techniques, like sequencing, that can only find point mutations. Moreover, unlike FISH, MLPA can detect small gene alterations.
- Results are available within 24 hours and because it is a multiplex reaction, it allows for a quick and efficient gathering of information.
- Small alterations to the MLPA protocol can allow for a variety of applications. For example, by adding an extra digestion step, MLPA can also be used to detect methylation patterns in DNA (Methylation specific-MLPA (MS-MLPA)).
Limitations of MLPA
- MLPA is extremely sensitive to impurities. Therefore, there needs to be extreme caution when preparing samples and performing the technique.
- There can be a decrease in signal from a probe due to a rare polymorphism or mutation, and it might be necessary to test it by other techniques.
MLPA is a great technique that can be used for varied applications and gives results quickly and efficiently. However, it doesn’t come without hiccups. As we all know, every technique has certain disadvantages, and each application needs to be thoroughly studied, to make sure we are using the most efficient and sensitive technique.
Have you ever used MLPA? What are your thoughts on this technique?
References
- Schouten, Jan P., et al. “Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification.” Nucleic Acids Res. 2002 Jun 15;30(12):e57.
- MRC-Holland. How Does MLPA® Work?