Today, the gut microbiome is garnering a large amount of media attention for its role in human health and disease. From influencing immune responses to impact our brain, the gut microbiome is an important and necessary aspect of our life. So much so, that current investigations in the gut microbiome are focusing on developing biomarkers for disease detection and therapeutic applications! For those who are interested in this field of research, here are 4 easy steps to get you started.
1. Define Your Study
Depending on your objectives, study types can be divided into three groups: longitudinal, cross-sectional or cohort, and intervention studies. If you want study the microbiome composition over time in the same person, a longitudinal study is the best choice. However, if you want to compare the microbiome composition between two different individuals, then a cross-sectional or cohort study is the right way to go. If you want to see the difference in the microbiomes of people who receive a treatment (e.g. drug, diet, and surgery) versus those who do not, then an intervention study is best.
2. Decide the Type of Samples and Collection Method
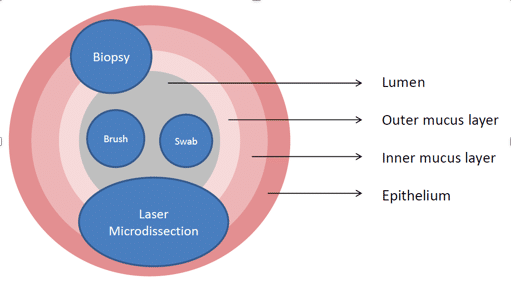
As we know, each type of sample offers different advantages and disadvantages. The classic good old-fashioned fecal sample is probably the most convenient. It is also the most versatile if you are planning to monitor microbiome changes frequently or execute a large-scale study. The non-invasive nature of this sample collection method directly translates into no bleeding and no bowel discomfort. Therefore, this method can help to reduce a subject’s dropout rate. However, the sample might contain dead bacteria. Also, you have no control over how sample is collected which leads to more sampling variables. The rectal swab is arguably easier to sample frequently compared to a fecal sample (Figure 1). However, there is a chance of getting skin bacterial contamination and inadequate biomass if not done properly.
The luminal brush is a better method when you want to assess host-microbe interactions. This is because you get a greater proportion of bacterial to host DNA, particularly in comparison with biopsies. It also results in greater mucosal coverage than biopsies (Figure 1). Despite this, there is a chance of obtaining lower biomass relative to other methods. A biopsy’s strong point lies in its ability to target an exact area of interest. Biopsies are probably the most painful for the subjects and not ideal in large study1.
Recently, a novel method of laser capture microdissection offers a way to directly isolate specific sections of complex tissues. It results in greater precision by fixing targeted cells to a certain platform with a laser beam2.
3. Determine a Suitable DNA and RNA Extraction Method
Generally, it is acceptable to store a sample at –20 °C within 15 min after defecation. However, this storage method is not applicable if you are working on a large study and have hundreds and thousands of samples. Instead, consider storing them at a lower temperature to prevent further damage to the DNA and RNA due to long waiting periods. A high proportion of unwanted host DNA and RNA can make bacterial nucleic acid extraction a complex task. The absence of a gold-standard method suitable for all samples only makes it worse. Therefore, it is important to consistently apply a single suitable extraction protocol. This will help to minimize the standard deviation throughout the study. If you are not sure about what method to use, I recommend you visit the International Human Microbiome Standards website.
4. Choose Your Sequencing Platform and Bioinformatic Pipeline
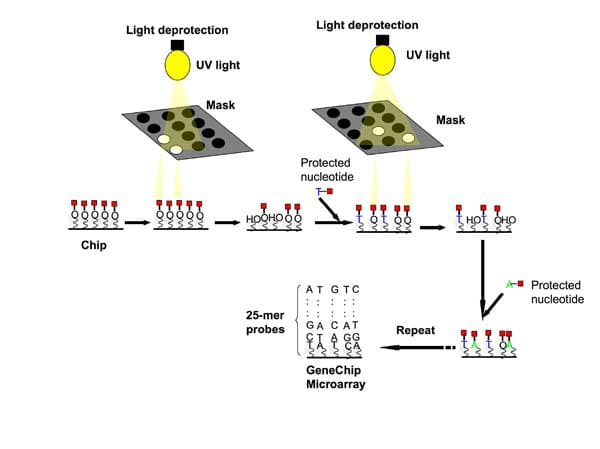
When sequencing an amplicon around 300 bp, most people use something like Illumina MiSeq and Ion PGM. By way of example, if you want to cover variable 16S rRNA sequences to create a phylogenetic tree, this platform will help to improve the accuracy of your analysis. As for shotgun sequencing, you might need more sequencing reads to cover the whole genome. The various analytical pipelines you get from the sequencing instruments are extremely limited in what they can do. Therefore, I recommended Mothur and QIIME to generate the basic analysis first if you are dealing with the 16S rRNA amplicon. For an advanced analysis of complex data from shotgun sequencing, you might want to team up with someone who can perform multivariate analysis.
That’s all for now! Do you have any questions or tips that I might have left out in this article? Feel free to comment below.
References:
- Claesson MJ, Clooney AG, O’Toole PW (2017). A clinician’s guide to microbiome analysis. Nature Reviews Gastroenterology & Hepatology 14:585-595.
- Zhang S, Cao X, Huang H (2017). Sampling strategies for three-dimensional spatial community structures in IBD microbiota research. Front Cell Infect Microbiol. 7:51.