Previously, I introduced the DNA microarray technology and described the principle behind it: hybridization between the nucleic acid sequence from the biological samples being examined and a synthetic probe immobilized and spatially arrayed on a solid surface, the microarray.
In this article, I will explain how these probes are designed and positioned on the array. I will also describe different kinds of DNA microarray platforms widely used for profiling gene expression.
So we know the sequence of genes we want to interrogate and we can design probes complementary to these sequences. The next task is to synthesize and attach these complementary probes in exact locations to create a high-density microarray. There are several possible approaches:
Spotted DNA Microarrays
The first DNA microarrays were spotted with probes that were made by oligonucleotide chemical synthesis and then attached to the array. These probes have to be “spotted” or “printed” using a robot onto a very fine grid by a sort of specialized inkjet-like printer, which uses the same technology as computer printers to expel nanoliter to picoliter volume droplets of probe solution, instead of ink, onto the slide.
Alternatively, these probes can be applied with a pin directly onto a specific location on the surface. The number of spots (aka features) applied onto the DNA microarray is limited to prevent cross-contamination problems.
In Situ-Synthesized DNA Microarrays
In situ-synthesized arrays are high-density oligonucleotide probe DNA microarrays, with Affymetrix GeneChip arrays being the most common. These are made using photolithography, which literally means to use light to create a pattern.
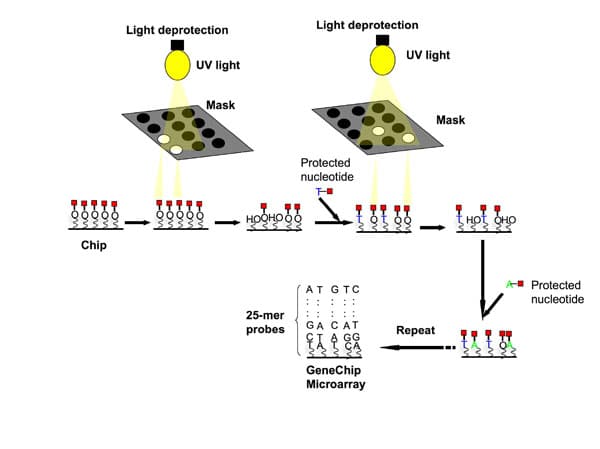
The method relies on UV masking and light-directed combinatorial chemical synthesis on a solid support to selectively synthesize probes directly on the surface of the array, one nucleotide at a time per spot, for many spots simultaneously (Figure 1).
This process works in the following way: a solid support contains covalent linker molecules that have a protecting group on the free end that can be removed by light. UV light is directed through a photolithographic mask to deprotect and activate selected sites with hydroxyl groups that initiate coupling with incoming protected nucleotides that attach to the activated sites.
The mask is designed in such a way that you can choose the exposure sites, and thus specify the coordinates on the array where each nucleotide will be attached. The process is repeated, a new mask is applied activating different sets of sites and coupling different bases, allowing arbitrary DNA probes to be constructed at each site.
This process is used to synthesize hundreds of thousands of different oligonucleotides. However, it is the length of oligos, not their number, that determines the number of steps required, since many different sites could be synthesized simultaneously.
Each probe on the chip requires four masks per round of synthesis: one mask to allow the addition of the required base and three other masks to prevent light from deprotecting the same spot while the other three nucleotides are being added. On average, each probe is 25 nucleotides long, requiring about 100 masks per chip!!!
These microarrays generally employ multiple probes for each gene to improve specificity and feature a match/mismatch probe pair that enables the discrimination of single mismatched base pairs.
Alternatives to the Affymetrix platform include Agilent arrays that use an inkjet spotting process for in-situ oligonucleotide synthesis, using five “ink” printing of the 4 nucleotide precursors plus catalyst, combined with coupling and deprotection steps that do not require the use of photolithographic masks.
This technology relies on printing picoliter volumes of nucleotides on the array surface in repeated rounds of base-by-base printing that extends the length of specific oligonucleotide probes.
This approach, therefore, allows the synthesis of longer molecules (60-mer length oligos) for their probes. Increased length improves the specificity of probes but at increased complexity of design, which reduces the number of features (Affymetrix chips usually feature >106 spots per microarray, compared to 0.24×106 features for Agilent).
High-Density Bead DNA microarrays
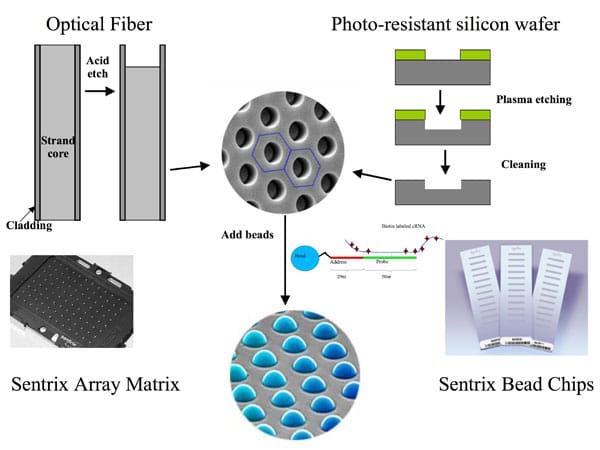
Another type of high-density DNA microarray are the BeadArrays manufactured by Illumina (Figure 2). Illumina’s Bead Array Technology is based on color-coded 3-micron silica beads that randomly self assemble in either a fiber-optic bundle substrate that then themselves assemble into arrays, or a silica slide substrate.
When randomly assembled on one of these two substrates, the beads have a uniform spacing of approximately 5.7 microns, with a packing density of about 40,000 array elements per square millimeter. This gives the Bead Array platform about 400 times the information density of a typical spotted array.
Each bead is covered with hundreds of thousands of copies of a specific oligonucleotide that act as the capture sequences in one of Illumina’s assays. Each bead has a 23-mer oligo “address” attached to it, which then anchors a 50-mer sequence-specific oligo probe.
The beads are randomly scattered across etched substrates during the array production process, with each array bundle containing about 50,000 beads. With this platform design, a specific oligonucleotide sequence is assigned to each bead type, but is replicated about 30 times on the array at random positions. Each gene is represented by two probe sequences.
A series of decoding hybridizations are used to determine which oligos are present at each matrix coordinate for every array.
These early DNA microarrays were instrumental for studying changes in steady-state levels after induction. However, transcriptional regulation of gene expression is only half the story. Steady-state measurements of transcript levels provide no information about changes in final protein expression that could be due to post-transcriptional regulation mechanisms. In addition, the microarrays described above are 3’-end biased because they rely on primers targeting the poly-A tail of mRNAs (at the 3’ end of the transcript), and contain probes that are clustered for detection of sequences at the 3’-end of the target.
While 3’ expression arrays (e.g. Affymetrix HG U133 plus 2 arrays) can report steady-state changes in transcript abundance, they are only able to distinguish transcripts of the same gene if the transcripts differ in their last few transcribed exons.
Many changes in transcript expression are caused by the inclusion of alternate exons, which would produce different isoforms of the gene, and thus would be invisible to 3’ traditional expression arrays. To address the challenge of alternative splicing that produces alternative exon expression, several splice-sensitive microarray platforms have been developed.
In the next instalment, I will introduce DNA microarrays designed specifically to detect exon expression in addition to transcript expression, and explain how alternative splicing analysis is performed. These arrays contain whole transcript preparation methods and “exon-centric” probes designed to circumvent the 3’-end bias and measure the overall abundance of the transcript as well as individual exons per transcript.
References
1. Miller MB, Tang YW (2009) Basic concepts of DNA microarrays and potential applications in clinical microbiology. Clin Microbiol Rev 22: 611-633.