We use fluorescent in situ hybridization (FISH) techniques routinely to detect DNA or RNA sequences in tissues, but what about micro RNAs (miRNAs)? No worries, FISH is now optimized to meet the challenge. To help you get going with the method, here’s what you need to know.
The first thing that comes to mind when we think of miRNAs is that they are small. Indeed they are short, single-stranded, and only 18-23 nucleotides long. Their size raises many concerns over the feasibility of FISH. Are they stable enough to avoid degradation in archival material? Can we make the probes sufficiently sensitive and specific? Will such small probes be lost by diffusion? What detection system is suitable for signals coming from such short targets? Let’s address these questions one by one.
The Integrity of miRNAs in FFPE Sections
Interestingly, studies show that miRNAs in formalin-fixed paraffin embedded (FFPE) tissues are even better preserved than mRNAs.1 Precisely because of their small size and close association with large protein complexes, they endure formalin fixation and paraffin embedding. Don’t relax too much though, as you need to try to keep an RNAse free environment at all times. Always use diethylpyrocarbonate (DEPC)-treated water for solution preparations. Also, autoclave everything that comes near your experiment!
Antigen Retrieval
Cross-linking during formalin fixation can limit probe and reagent access to target miRNAs. You can unmask these sites with several antigen retrieval procedures. Most protocols use proteinase K in Tris buffer to break the formaldehyde crosslinks. Others report that proteinase K may destroy some of the epitopes and advise heating the specimens in citrate buffer instead. As both approaches work, just check which procedure your slides like better.
Probes
The first ventures in FISH miRNAs tried digoxigenin labeled ribonucleotide probes. However, due to the short length of the micro RNA target, the duplex was not sufficiently stable. To make things worse, high sequence similarity between closely related miRNAs made it difficult to achieve sufficient specificity. Today we mostly use locked DNA (LNA) probes which perform way better.
Locked DNA is a class of DNA analogs in which the sugar ring is locked with a methylene bridge stabilizing the probe in an N conformation. LNA probes are thus optimally preorganized for binding. They achieve higher affinity binding and a more stable duplex at much higher temperatures. Mismatch discrimination is greatly improved, as greater temperature difference exists between a perfectly matched duplex and a single mismatch duplex. This makes it much easier to find a hybridization temperature at which only the perfect match is detected. Finally, the LNA/miRNA hybrid is resistant to nuclease attack.
Control Probes
Make sure to use both positive and negative control probes, especially while setting up the procedure. For a positive control you should detect a miRNA that is specific for your tissue. As for negative controls, a probe with two mismatches and a probe specific for a miRNA that is known not to be expressed in your tissue are advised. In case you are having trouble distinguishing the signal from the background noise, you should also include a ‘no probe’ experiment.
Hybridization and Stringency Wash
The best thing is to prehybridize your sections in prehybridization solution at 50°C for 2h, and then hybridize with the probe overnight, also at 50°C. Then you perform stringency washes, which are crucial for the removal of non-specific interactions. Do your stringency washes in SSC buffer, first at 37°C, followed by 50°C with shaking. The first wash washes away the excess probe and hybridization buffer, while the second one removes the non-specific and repetitive hybridization. If non-specific binding is still an issue, try repeating the washes several times. If you are just getting started, you can play around a bit with the hybridization and the stringency temperatures. Sometimes two degrees up or down can make a huge difference.
Probe Labeling and Signal Detection
Nowadays, you can choose from a wide palette of commercially available tracers for oligonucleotide labeling. Both biotin and dioxigenin are useful as 5’conjugates, suitable for detection by immuno-histochemistry. In case your tissue is rich with endogenous biotin, dioxigenin should definitely be your first choice. After binding the anti-label antibody, use a horse radish peroxidase (HRP)-based signal amplification system to visualize the reaction.
You can also choose an appropriate fluorophore for your HRP substrate. Most commonly in use are the cyanine dyes, but Alexa series or Quasar dyes are also an option. The choice is yours. The decision primarily depends on required ex/em wavelengths, possible double-labeling, and, of course, availability and funds.
Oh the Possibilities…
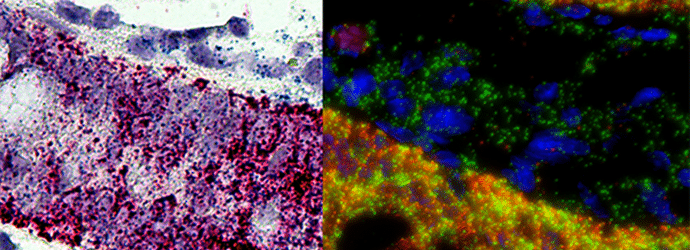
The advantages of detecting miRNAs by FISH in FFPE are obvious. You can visualize their exact location in complex tissues. You can go through a large number of samples and examine them retrospectively. Now there are protocols to simultaneously detect miRNAs with protein markers2, as well as methods for multiplex miRNA FISH.3 Bear in mind that miRNA research is gaining momentum, both in cancer research and in developmental biology. So explore your possibilities!
References
- A. Liu, M. T. Tetzlaff, P. VanBelle et al. (2009) MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. International Journal of Clinical and Experimental Pathology, 2, (6), pp. 519–527.
- A. Chaudhuri, S. Yelamanchili and H. Fox (2013). Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Frontiers in Cellular Neuroscience, 160 (7), pp 1-8
- N. Renwick, P. Cekan, C. Bognanni et al. (2014) Multiplexed miRNA Fluorescence In Situ Hybridization for Formalin-Fixed Paraffin-Embedded Tissues. Methods in Molecular Biology, 1211, pp:171-187