In part 1 of The Who’s Who of Super Resolution Microscopy I discussed one way to overcome the diffraction limit of light: ensemble technique. In this article I discuss another way to improve resolution: single particle localisation techniques.
Single particle localisation techniques works by pin-pointing single molecules by reconstructing a super-resolution image from multiple frames (usually hundreds!). The concept behind the single molecule localisation techniques is that a single fluorophore, even though it is subject to the same diffraction limit as confocal microscopy, can be precisely localised by calculating the centroid (central point) of the point spread function or PSF (see part 1 for a re-cap!). This is usually done by subjecting the resulting image to mathematical fitting such as fitting to a Gaussian distribution and then localising the central point of each spot. We know that not all of you are confident in using mathematical fittings in your data, but that’s ok! It is enough to understand the basics. But if you want to know more about this idea, check out Thompson et al, 2002.
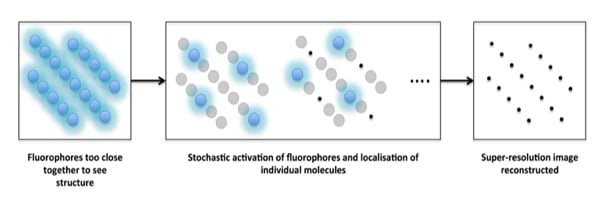
But how do we get this from theory to isolating the each individual fluorophore from the blur which results from the large PSFs? This is where it gets clever! By keeping the activating laser beam at a low power, the fluorophores are excited ‘stochastically’, meaning that only a handful of all the fluorophores in a given image are excited in each frame. These fluorophores are imaged until they are bleached, and the cycle is repeated as you collect more and more frames. By thus acquiring a large number of frames – and then localising the centre of the PSF of each particle detected within each frame – a full super-resolution image can be reconstructed (Figure 1).
So rather than altering the PSF, as in STED microscopy, single molecule localisation techniques achieve super-resolution by extensive post-acquisition image processing.
So how is this done?
The two main single molecule localisation techniquesare PALM and STORM, which were invented around the same time. They are both based on the idea outlined above, but differ in the use of fluorophores.
Single Molecule Localisation Technique #1: PALM (Photoactivated localisation microscopy)
The technical stuff
PALM uses photoswitchable and photoactivatable fluorescent proteins. In both of these cases, exposure to a specific wavelength causes the fluorophore to change its emission spectra – either from a non-fluorescent to fluorescent state (photoactivatable) or from one fluorescent state to another (photoswitchable). These photoswitchable and photoactivatable fluorescent proteins can be tagged onto your protein-of-interest in the same way as you would tag a protein with GFP.
How does this help you?
The potential for genetically-encoded labelling is one of the main advantages of this method, as you do not have to worry about non-specific binding and it is compatible with live cell imaging. The disadvantages are that you get lower photon counts with fluorescent proteins than with labelling with a dye, so it may be harder to obtain the full super resolution image.
Examples of how it has been used
- PALM has been used to show the presence and reorganisation of nanometre-sized clusters of NK cell receptors at the immune synapse (Pageon, et al 2013)
- Live cell PALM has been used to image the adaptor protein Lat at the T cell synapse (Williamson et al, 2011)
Single Molecule Localisation Techniques #2: STORM (Stochastic optical reconstruction microscopy)
The technical stuff
STORM as primarily implemented using photoswitchable fluorescent dyes coupled to antibodies. This means that it is limited to fixed cell imaging. Direct STORM (dSTORM) is a variant of STORM based on the reversible photoswitching of conventional fluorophores into a long-lived ‘dark state’ (an energy level where the molecule cannot emit photons). This allows the STORM technique to use more conventional dyes such as AlexaFluor488 and 647, which makes experiment design and sample prep easier.
How does this help you?
The advantage of STORM over PALM is that you have control over antibody concentrations, and may be able to get a brighter stain and therefore better imaging processing. Also, using antibodies means you do not have to clone and transfect your protein-of-interest. This is a real plus if you need to image whole tissues – if you can avoid genetically engineered mice by all means do. However, STORM is vulnerable to background in the form of non-specific antibody binding.
Examples of how it has been used
- STORM has been used to study the transport dynamics of cargo along microtubules (Balint et al, 2012)
- 3D STORM has been used to look at the three dimensional filament organisation of the actin cytoskeleton (Xu et al., 2012)
All of the super resolution microscopy techniques that I covered in these last two articles have the potential to make big discoveries in the field of biology. So if you are lucky enough to have one (or two!) of these microscopes at your disposal get to using them!
I hope this has given you a good (albeit basic) overview of how super resolution microscopes work and what you can do with them. Keep your eyes open for my future articles on other super-resolution themes, including what to do with the all important data once you have it!
References
Balint, S., Vilanova, I., Alvarez, A. and Lakadamyali, M. (2012) Correlative live-cell and Superresolution microscopy reveals cargo transport dynamics at microtubule intersections. PNAS 110:(9);3375–80.
Pageon, S., Cordoba, S., Owen, D., Rothery, S., Oszmiana, A. and Davis, D. (2013) Superresolution microscopy reveals nanometer-scale reorganisation of inhibitory natural killer cell receptors upon activation of NK cell. Science Signalling, 6:(285);ra62.
Thompson, R., Larson, D. and Webb, W. (2002) Precise nanometer localisation analysis for individual fluorescent probes. Biophysical Journal 82:(5);2775–83.
Williamson, D., Owen, D., Rossym J., Magenau, A., Wehrmann, M., Gooding, J. and Gaus, K. (2011) Pre-existing clusters of the adaptor Lat do not participate in early T cell signalling events. Nature Immunology 12: 655–62.
Xu, K., Babcock, H. and Zhuang, X. (2012) Dual-objective STORM reveals three-dimensional filament organisation in the actin cytoskeleton. Nature Methods, 9: 185–188.