Ligation independent cloning (LIC) is an easy and effective method to ensure successful cloning, all without the need for ligation. As easy as the technique is, designing primers can be a bit tricky. In this article, we will present a quick overview on primer design for ligation independent cloning.
The easiest way to start is to look at the treated vector that the insert will be annealed into. In the diagram below, panel 1 shows the treated vector waiting for the insert and panel 2 shows the corresponding insert, which would fit into it. It’s easy to see how those would fit together, but how do you make the insert like that?
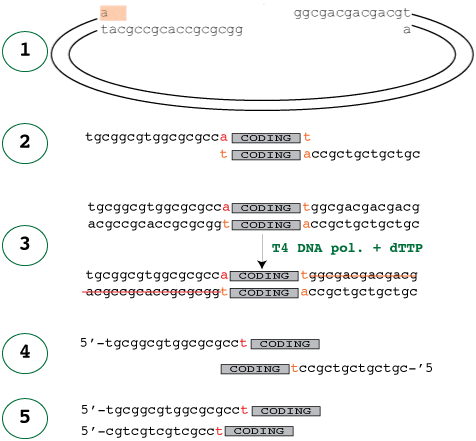
Just like in the vector treatment, the 3′-5′ exonuclease activity of the polymerase digests the 3′ ends of the insert because no are dNTP’s available. This activity stops once it reaches dTTP, where the polymerase and exonuclease activities cancel each other out, resulting in stable, long sticky ends.
It is crucial to note that the forward primer is designed so that the addition of the extra T does not disrupt the reading frame of the N-terminal fusion leading up to the coding sequence.
Panel 4 shows the primers required to amplify the insert with the ligation independent cloning vectors. Of course, one of the primer sequences – the reverse primer – has to reversed to that it reads 5-3′ before it is sent off for sequencing. A common error is to reverse complement this sequence, which of course is wrong – it only needs to be reversed as it is already the complement.
The good news is that you only have to design these adaptor parts of the primers once. As long as you always use the same ligation independent cloning site, the adaptor parts will always should be the same. The parts of the primer that are specific to the gene to be amplified should be designed as normal, and the adaptors simply added to the ends. Note also that since the start codon is vector-borne, the start codon of the gene itself should be omitted.
If you are still confused (you may well be), the best thing to do is try it yourself – get a pencil and a piece of paper, and sketch out the process to become more comfortable with understanding how it works.
You may also find this tool useful for helping to design LIC primers.




