Nanopore Sequencing: An Update
People would have said that a USB sequencer not much bigger than a memory stick which could sequence genomes in 50kb+ read-lengths was impossible “’Star Trek’ technology!” Now, that futuristic technology is here.
Join Us
Sign up for our feature-packed newsletter today to ensure you get the latest expert help and advice to level up your lab work.


People would have said that a USB sequencer not much bigger than a memory stick which could sequence genomes in 50kb+ read-lengths was impossible “’Star Trek’ technology!” Now, that futuristic technology is here.
RNA sequencing (Wang 2009) is rapidly replacing gene expression microarrays in many labs. RNA-seq lets you quantify, discover and profile RNAs. For this technique, mRNA (and other RNAs) are first converted to cDNA. The cDNA is then used as the input for a next-generation sequencing library preparation. In this article, I’ll give a brief…
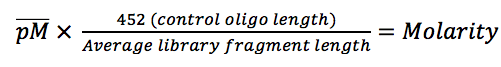
If you want to get the maximum yield and quality from your next-generation sequencing experiment then you are going to need to make sure each of the libraries you produce is carefully quantified ready for pooling and/or loading onto a flow cell. If the quantification goes wrong you’ll get a bad balance of samples within…

Making a Next Generation Sequencing (NGS) library can seem a bit daunting to the new user, as failures can be expensive. But don’t be put off, as NGS library preparation is relatively simple molecular biology, and can be very easy if you choose to use a commercial kit from one of the many suppliers. Take…
Next-generation sequencing (NGS) really has taken the world by storm! In NGS, millions of short ‘read’s are sequenced in a short space of time, leaving you with vast amounts of data to analyze! For all NGS platforms, the input sample (i.e. your cell free DNA) must be cleaved into short sections or fragments prior to…
Size Selection via Gel Electrophoresis Whether you are using NGS for whole genome sequencing, SNP variant analysis, HLA typing, HLA matching, or even transcriptome or miRNA analysis by RNA-seq, size selection is an extremely important consideration for optimum results. Precise size selection can increase sequencing efficiency, save money and improve genome assemblies, as well as…
You’ve carefully collected your samples, extracted nucleic acids and made your first set of next-generation sequencing libraries. How are you going to know if the data you get back is any good and whether it will be worth the effort in learning how to do the analysis? Who is to blame? Fortunately, there are several…
Sanger sequencing is still a workhorse of most molecular biology labs. Even with the advent of next-generation sequencing we still need to sequence our clones and PCR products. In this article I have listed some of the tips and tricks we used in our Sanger services. (1)Dilution of BigDye: I’d expect this to be a…
The Human Genome Project was successful, but hard work. The major improvements to the technology were the increases in parallelization and automation. In 2003, just as the HGP completion papers were published in Nature and Science, ABI launched the‘3730XL’. It could run 24 96-well plates per day and generate around 2 MB of sequence. Some…
It all started with proteins The earliest methods for sequencing were developed for proteins. In 1950, Pehr Edman published a paper demonstrating a label-cleavage method for protein sequencing which was later termed “Edman degradation”. Around the same time Fred Sanger was developing his own labelling and separation method which led to the sequencing of insulin….
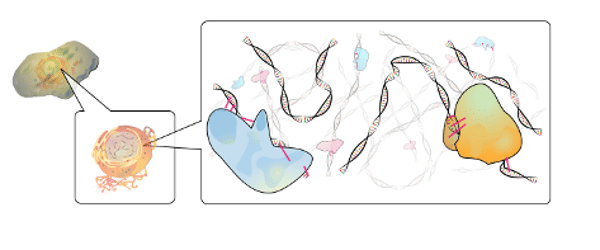
ChIP-seq is a wonderful technique that allows us to interrogate the physical binding interactions between protein and DNA using next-generation sequencing. In this article, I’ll give a brief review of ChIP and introduce the chromatin immunoprecipitation sequencing technique (ChIP-seq), which combines ChIP with next-generation sequencing. What is chromatin immunoprecipitation? Chromatin immunoprecipitation (ChIP) allows us to…

The eBook with top tips from our Researcher community.