You’ve carefully collected your samples, extracted nucleic acids and made your first set of next-generation sequencing libraries. How are you going to know if the data you get back is any good and whether it will be worth the effort in learning how to do the analysis?
Who is to blame?
Fortunately, there are several quality controls you can look at to see if your sequence data are high-quality. Issues arising are usually due to one of these three; your library, the sequencing or the sequencing lab. As per usual, my focus is on Illumina sequencing, but similar metrics are available from other sequencers. You should consider any individual QC metric in context, failure for one run might be a pass for another, and different library types will give different metrics. Use your QC analysis as a starting point for investigating if something has gone wrong and why.
Did it actually run?
Probably your first question will be “did my run work and are there enough sequences?” If the run obviously failed hopefully you never got the data, and if there were not enough sequences hopefully your service provider repeated the run and gave you some more! A rule of thumb is that you should see over 10 M reads from a MiSeq run, >20 M reads from a GAIIx lane and >100 M reads from a HiSeq lane. Furthermore, 70% of the data should be Q30 or higher.
Don’t stop me now
Don’t stop there though as the tools available to you are very useful, and although there are too many to list them in this article, I have chosen to focus on three of the most useful for checking Illumina data.
(1) RTA and SAV: These are Illumina tools used to monitor sequencing during or after the run and your service provider may send these files to you along with your data. The most significant metrics in Illumina’s ‘Sequence Analysis Viewer’ (SAV) reports are:
- Number of reads- did you get >100 M from your HiSeq lane?
- Percentage >Q30- is your data >70% Q30?
- Error rate- is the error rate <0.5%? A word of warning- this metric is very dependent on the length of your read and where you measure the error rate.
- Demultiplexing- are your barcoded and multiplexed libraries well balanced or are there libraries under- or over-represented?
(2) FastQC: This very useful and relatively easy to use tool was developed by Simon Andrews at the Babraham Institute in Cambridge, see the FastQC homepage. It allows you to import your sequence data in the common formats, BAM, SAM or FastQ and get a quick overview of sequence quality. Each metric is reported with a traffic light warning system, normal (green), abnormal (orange) or bad (red), in a static web page.

The most useful plots for users are:
- Per Base Sequence Quality. This plots the Qscore of the raw sequence reads as a box-plot for each cycle. Higher is always better, and the characteristic decay of quality is seen in most runs.
- Per Base Sequence Content. This plots the proportion of each base at each cycle. In a random fragment library from a ‘normal’ genome you would expect to see all four bases equally represented. Deviation from normal base content can indicate issues with library quality, but equally some genomes are very GC biased and some NGS applications also introduce a strong GC bias (such as Bis-seq).
- Duplicate Sequences. This plots the number of times the same sequence is seen in a 200,000 read subset of your data. In most libraries you would hope to see >10% duplicate rates. If this number is high in your library it can indicate over-amplification or poor library-prep.
(3) MiSeq QC: While not strictly a tool to check data from your own genome project, the MiSeq system does offer the option of performing a QC run on your libraries before you get them deep-sequenced on a HiSeq.
The MiSeq QC run is a quick paired-end 50 bp run. This generates enough data for relatively high-quality alignment and will allow you to get a good idea of how well your libraries will perform on HiSeq. The same metrics are reported in SAV (see above) and this is available in ‘BaseSpace’, lllumina’s cloud-based analysis environment.
Check before you spend
You can also run the data through FastQC and hopefully this will appear as an ‘App’ in BaseSpace at some point. The real benefit of the MiSeq QC is that it gives a real sequence based quality check of your whole experimental process before you ask for a whole HiSeq flowcell which might cost £5 to £10,000 or more.
A robust assessment
Even if you have 96 multiplexed libraries in a pool, you should still get over 100,000 reads from each library allowing a robust assessment of quality, duplication rate and barcode-balance. We are using MiSeq QC for each plate of samples we run through the library-prep services in my lab.
Nasty Surprise!
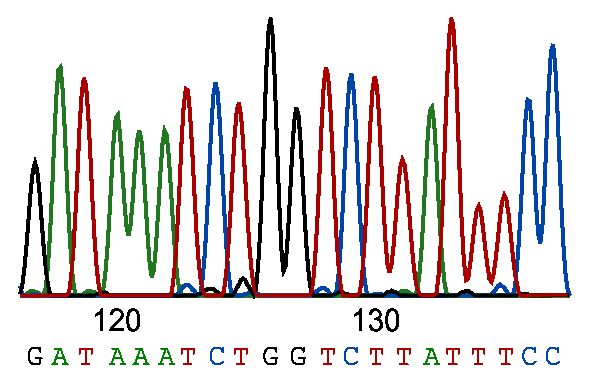
Hopefully your QC check does not throw up any nasty surprises. If it does then take the time to use these tools in discussion with whoever did the sequencing and try to find out what went wrong. Similarly to Sanger sequencing, the template is often the cause of failure. Bad samples rarely make good libraries for sequencing.