Do you want to start using the CRISPR-Cas13 system in your experiments? Then you will need to know how to design a Cas13 gRNA.
Cas13 is a CRISPR-Cas protein that can help you to study and manipulate RNA in mammalian cells. For Cas13 to work, it needs to find the RNA you want to modify. Targeting your desired RNA is easily achieved with the help of a short RNA. Because it guides Cas13 to your target, this RNA is called a guide RNA (gRNA).
This article explains how to design gRNA for the transcript you want to target and provides a checklist to ensure your experiments run smoothly.
What is a gRNA?
The CRISPR system has two main components: a Cas protein (Cas 9, 12, or 13) and the gRNA. The Cas protein has nuclease activity, and the gRNA leads the Cas protein to the specific target. A gRNA for Cas13 is a small RNA with a conserved sequence forming a stem-loop structure (also known as direct repeat or ‘DR’) and a spacer sequence. Cas13 binds to the DR, and the spacer sequence is complementary to the target nucleic acid. (Figure 1)
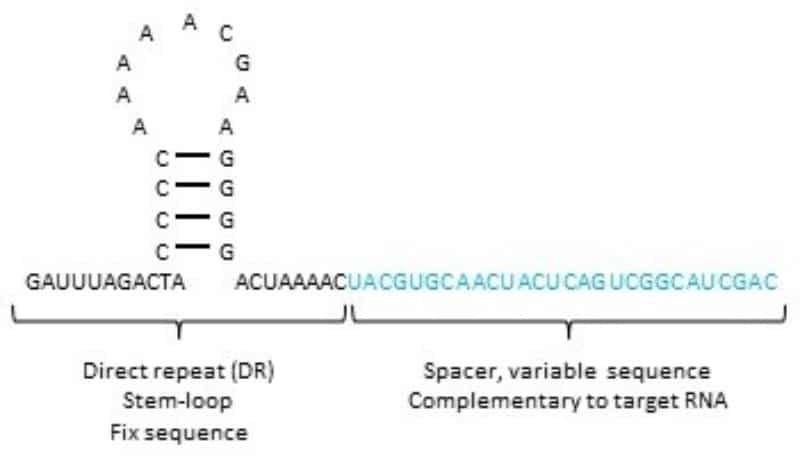
How Does Cas13 gRNA Design Differ from Cas9?
Cas9 requires two RNAs:
- the trans-activating CRISPR RNA (tracr-RNA); and
- the CRISPR-RNA (crRNA).
However, Cas13 only needs a shorter single RNA.
Additionally, unlike Cas9, Cas13 does not need a proto-adjacent motif (PAM) next to the target sequence to cut it. Some Cas13 proteins prefer an adenine, uracil, or cytosine next to the 3’ protospacer flanking site (PFS), but the PFS is not essential for Cas13 nuclease activity. [1] This lack of requirement for a PAM makes targeting RNA a bit easier.
In theory, you could target any region of the RNA. However, Cas13 proteins only bind and cut ssRNA. mRNAs are single-stranded nucleic acids, but RNAs have a secondary structure that can generate loops and dsRNA. It is essential that your gRNAs for Cas13 target the loops or single-stranded regions of the RNA. [2]
Where Do I Start With Designing Cas13 gRNA?
The first thing you need to do is choose which Cas13 protein you will work with. It could be Cas13 a, b, c, d, etc. Each Cas13 protein has its advantages and disadvantages. Let’s say you want to use Cas13a. Now you need to check which bacteria strain the Cas13a you have comes from. Is it Leptotichia buccalis (Lbu), Leptotrichia wadei (Lwa), or another one?
This first step is crucial for designing gRNAs because it will determine the sequence of the DR you need. Cas13a binds to a different DR than Cas13 b, c, or d. Also, Cas13a proteins from other bacteria strains would only recognize their specific DR, and you would not want to use LbuCas13a with a gRNA for LwaCas13a because it will not work. See Table 1 below for a list of the DRs for the most commonly used Cas13 proteins.
Table 1: Characteristics of commonly used Cas13 proteins.
Cas protein | Species | DR Sequence | Reference |
LwCas13a | Leptotrichia wadei | GATTTAGACTACCCCAAAAACGAAGGGGACTAAAAC | [3] |
PspCas13b | Prevotella sp. | GTTGTGGAAGGTCCAGTTTTGAGGGGCTATTACAAC | [4] |
RxCas13d | Ruminococcus flavefaciens XPD3002 | CCCCTACCAACTGGTCGGGGTTT | [5] |
Let’s assume you are using LwaCas13a. Now that you’ve got the DR sequence, it is time to design the spacer sequence. This spacer sequence is the RNA that will tell Cas13a which RNA to target is a 22-28 nucleotide long sequence, complementary to the target sequence. Let’s discuss how to select which region of your RNA you should target.
How to Design the Spacer Sequence
As mentioned previously, Cas13 proteins do not require a PAM sequence, making the design stage simpler. Let’s walk you through this step by step.
Step 1: Get the sequence of your target RNA
To get the mRNA sequence, go to the NIH website and look for your gene of interest. In this example, we’ll pick hnRNPU. Find the mRNA sequence in the NCBI reference sequences section, and download the FASTA sequence (Figure 2).
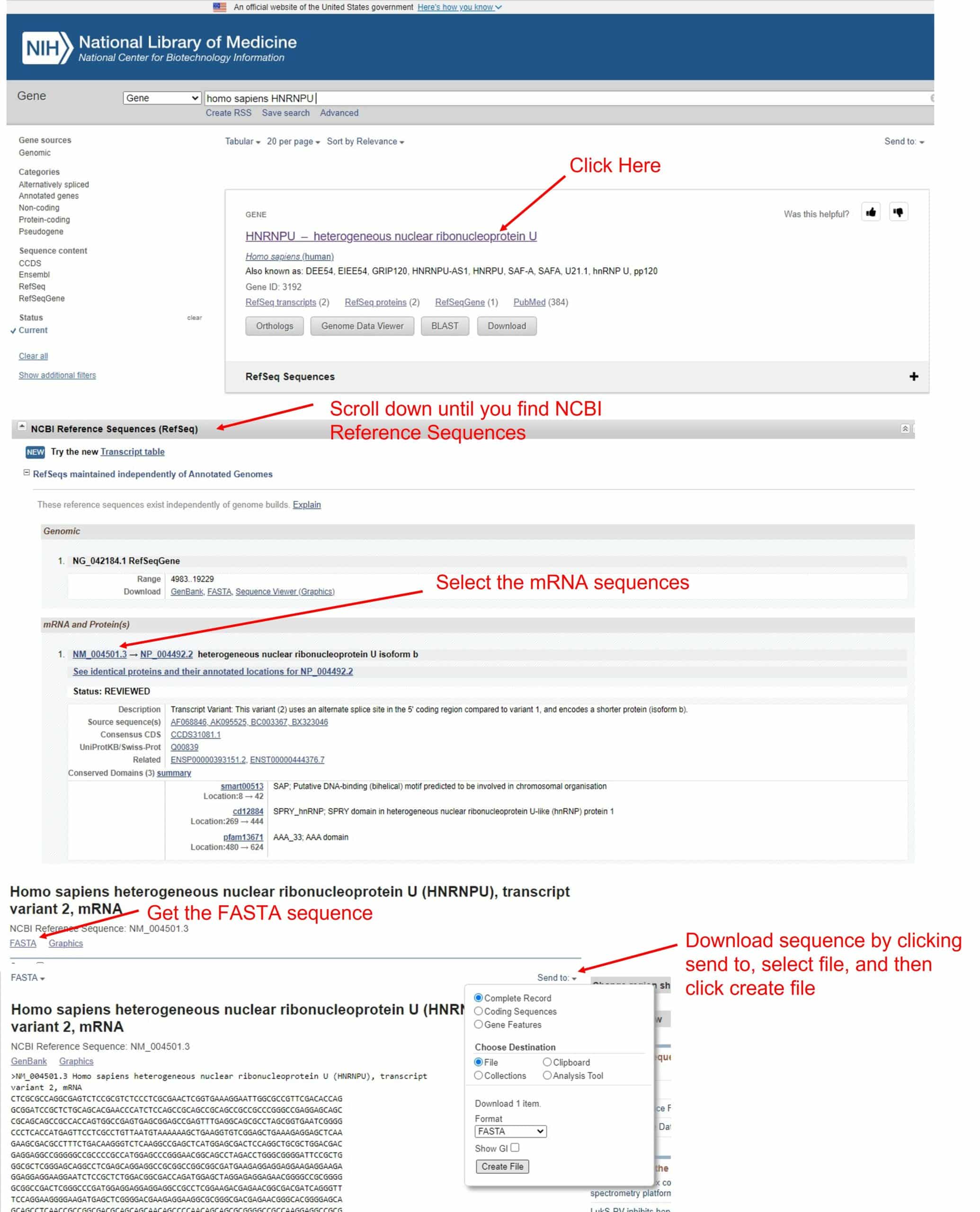
Step 2: Select the target sequence
You can use the Sanjana Lab online tool to select the region of your RNA that Cas13 will target. This tool follows the rules of the works of Wessels and Guo. [6,7] (Figure 3)

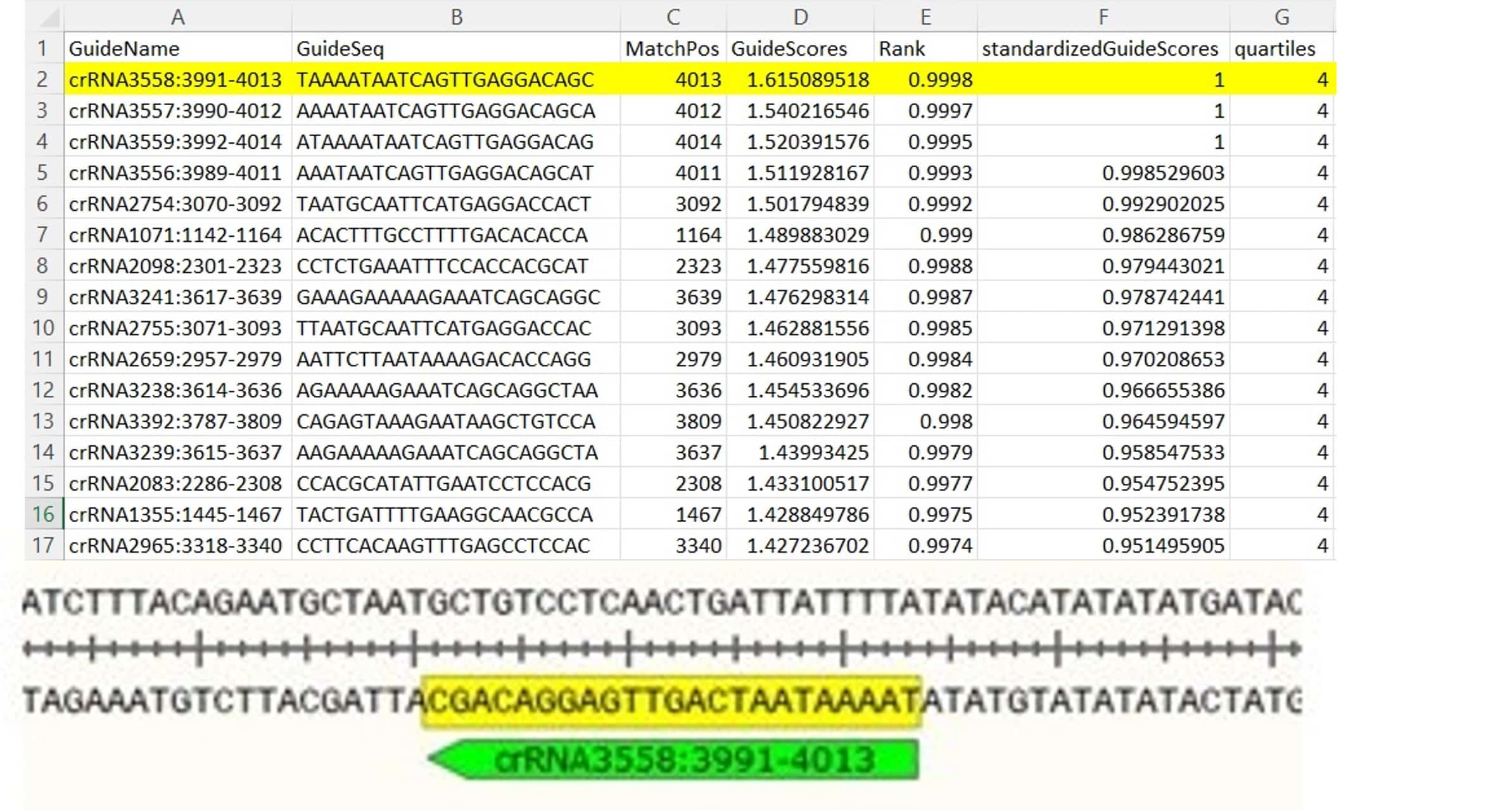
These rules include looking at the structure of the RNA and the effect of single nucleotide mismatches. It is friendly and easy to use. Select the design custom gRNAs option. Upload your sequence as a FASTA file, or paste it if it is shorter than 500 bases. Click the “design guides” button, wait a few minutes, and a table with the predicted gRNAs will be generated or sent to you by email.
This table gives an ID to each crRNA, the sequence, the RNA region the crRNA targets, a predicted efficacy score, quartile value (Q4 means the predicted efficacy is high, whereas Q1 would have a low predicted efficacy), and potential off-targets. Choose gRNAs with the highest quartile value (which is 4) and guide score. (Figure 4)
Step 3: Consider Isoforms
If your gene of interest is expressed as different isoforms, check that the gRNAs you selected target all of the endogenous transcripts. You can use BLAST to do this.
Step 4: Generate your gRNAs
You can have the gRNAs synthesized by a company or clone the crRNA in a plasmid.
You can find plasmids to clone your crRNA into in AddGene by searching Cas13 gRNA. There you can select the plasmid for bacteria or mammalian expression.
For mammalian expression, the plasmid has a U6 promoter, the DR specific for each Cas13 protein, a restriction enzyme site to allow insertion of the crRNA, and a transcription termination site.
If you cannot find the right plasmid on AddGene, you can regenerate it and submit it to the repository for others to use after testing it. Find out how to do it here.
Once your gRNAs/plasmids are ready, deliver them with Cas13 to your preferred cell line.
How to Test Your gRNAs
It is always good to test at least three gRNAs. Testing can be done by measuring the RNA levels of the transcript of your interest 48–72 hours after transfecting cells with Cas13 and the gRNA. RT-qPCR is the best and easiest way to do it. If your gRNAs work, you will see a reduction in the transcript levels. For future experiments, use the gRNA that gives the highest reduction in levels.
Things Worth Double-checking to Prevent Experimental Disaster
- The DR in your gRNA is the correct one for the Cas13 protein you use.
- The spacer sequence of your gRNA is complementary to your target RNA.
- You have designed three different gRNAs to test.
- A non-targeting gRNA control is included in your experiments.
Summary
The lack of PAM requirement by Cas13 makes it easier to design gRNAs for Cas13 than for Cas9. However, other factors, like the structure of the target RNA, need to be considered. Although there are few web tools to design gRNA for Cas13, the available ones are excellent and can be used for any Cas13 protein.
Whether you want to know how to get started with CRISPR or find new applications of this genome editing tool, head to the Bitesize Bio CRISPR Research Hub.
References
- Abudayyeh, O. et al. (2016) C2c2 is a single component programmable RNA-guided RNA-targeting CRISPR effector. Science. 353(6299):aaf5573
- Bandaru S. et al. (2020) Structure-based design of gRNA for Cas13. Sci Rep. 10;11610.
- Abudayyeh, O. (2017) RNA targeting with CRISPR-Cas13. Nature. 550(7675);280–284.
- Cox, D. et al. (2017) RNA editing with CRISPR-Cas13. Science, 358;1019–1027
- Konermann, S, et al. (2018) Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. pii: S0092-8674(18)30207-1.
- Wessels, HH. et al. (2020) Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol. 38;722–727.
- Guo X. et al. (2021) Transcriptome-wide Cas13 guide RNA design for model organisms and viral RNA pathogens. Cell Genomics. 1;100001.



