Has your research ground to a halt because you’re stuck trying to express an insoluble protein? Can you express and purify this protein but can’t demonstrate its activity?
I have two words that may help you: osmotic shock. Read on to find out how this technique can get you back on the road to successful protein research.
What Is Osmotic Shock?
Exposing bacterial cells to high external concentrations of osmolytes (small, soluble compounds that regulate cell volume) retards cell growth and division. This is because the cells lose water to the growth medium through osmosis, resulting in so-called osmotic shock. [1]
The shock isn’t lethal, and the cells eventually equilibrate to it by synthesizing and absorbing their own osmolytes. This prevents water loss and allows the cells to continue to grow and divide (Figure 1). In E. coli, the most common host for protein expression, these osmolytes include potassium, glutamate, and trehalose.
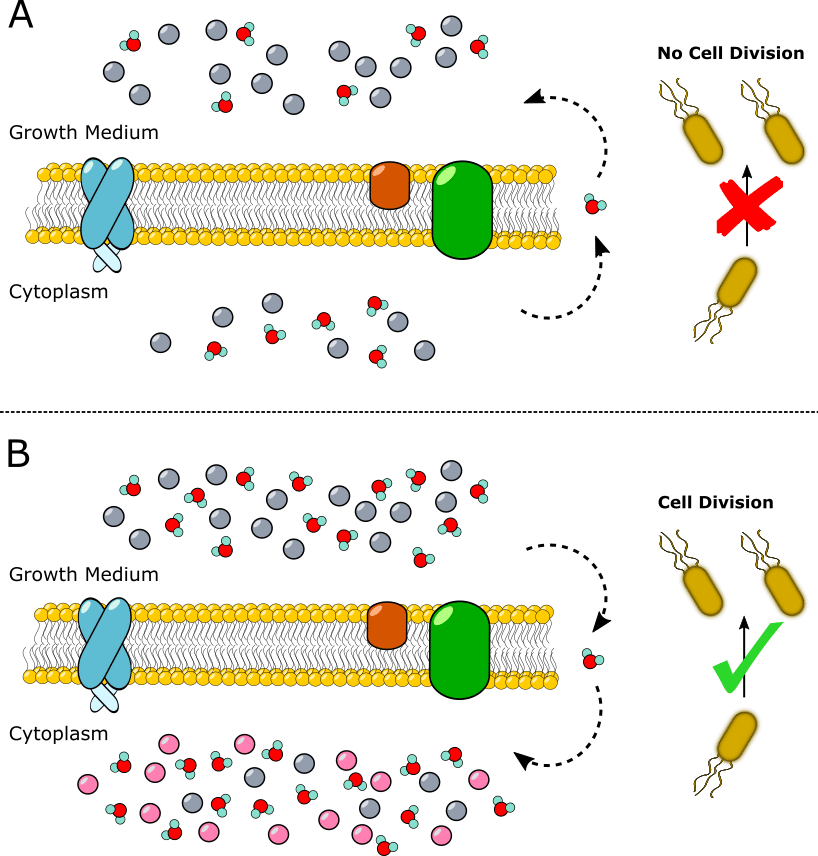
How Does Osmotic Shock Increase the Solubility and Activity of Expressed Proteins?
In short, nobody knows for sure. The theory with the most evidence going for it is the so-called osmophobic theory. [2] It asserts that chemical interactions between the osmolyte and the target protein backbone are energetically unfavorable. These interactions are presumed to be less prevalent when the protein is folded, so the native and presumably active form is promoted in vivo during expression.
Back to Reality: How to Perform Osmotic Shock
This is all well and good, but how do you apply this technique?
Well, it’s simple. Add 500–1000 mM D-sorbitol to your bacterial growth media, and proceed as usual. [3] Oh, and don’t worry about dissolving all that powder, it will magically disappear in the autoclave.
Another study reported that including 2.5 mM glycyl betaine in addition to the D-sorbitol further improves results, giving a 427-fold increase in activity for the test sample, so you could try that too. [4]
You could vary the NaCl concentration of your chosen medium as well, as NaCl is itself an osmolyte. Benchmark any changes in expression yield and target activity against a control expression and pow! Hopefully, you get some excellent results.
There’s Always a Downside
There is a drawback with this little gem of a technique. In synthesizing and absorbing extra osmolytes, the cells experience a significant metabolic burden. As a result, they grow and divide slowly. Really. Really. Slowly. Because of this, you will be waiting a long time for your bacterial cultures to reach an optical density at which it is appropriate to induce the expression of your target protein.
You can overcome this to some degree by including the osmolytes in your small-volume culture that you reinoculate into the large-volume cultures before expression. Since the small-volume culture is usually left overnight, the cells can equilibrate to the osmotic shock conditions while you’re happily sleeping.
If you’re new to all this protein expression stuff and these terms are a bit confusing, be sure to check out this excellent article on optical density and this guide to protein expression.
Even so, be prepared to wait two or three times as long as you would in the absence of osmolytes, and even longer if you are selecting for multiple plasmids with multiple antibiotics.
Did osmotic shock work for you? Discovered any additional osmolytes that helped your protein express? Be sure to leave a comment below if so.
And take your protein purification game to the next level with our two free eBooks: The Bitesize Bio Guide to Protein Expression and Five Methods for Assessing Protein Purity and Quality.
References
- Yancey PH (2001) Water Stress, Osmolytes and Proteins. Am Zool 41:699–709
- Bolen DW and Baskakov IV (2001) The osmophobic effect: natural selection of a thermodynamic force in protein folding. J Mol Biol 310:955–63
- Oganesyan N et al. (2007) Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization. Protein Expr Purif 52:280–5
- Blackwell JR and Horgan R (1991) A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett 295:10–2







